Chikungunya virus-specific CD4+ T cells are associated with chronic chikungunya viral arthritic disease in humans
- PMID: 40398392
- PMCID: PMC12147901
- DOI: 10.1016/j.xcrm.2025.102134
Chikungunya virus-specific CD4+ T cells are associated with chronic chikungunya viral arthritic disease in humans
Abstract
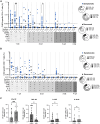
Chikungunya virus (CHIKV) is a mosquito-borne virus that can cause chronic chikungunya virus disease (CHIKVD), which is characterized by persistent incapacitating arthralgia. Despite recurring CHIKV outbreaks and recent approval of a vaccine, the breadth and target of T cell responses in CHIKVD remain largely understudied. Here, we tested peripheral blood mononuclear cells (PBMCs) collected from CHIKV-infected individuals against overlapping peptide pools sequentially spanning the entire CHIKV proteome. We detected robust CHIKV-specific CD4+, but not CD8+, T cell responses in infected individuals. Individuals with chronic arthralgia displayed significantly higher CD4+ T cell responses against nsP1, nsP2, and E2 proteins and exhibited a significantly lower Th1 CD4+ T cell population, compared to individuals who had recovered. Additionally, CD4+ T cells in chronic individuals were marked by a predominant production of tumor necrosis factor alpha (TNF-α). Overall, our work comprehensively characterizes T cell responses in CHIKVD in humans and provides insights into the role of T cells in CHIKVD.
Keywords: CD4(+) T cells; T cells; chikungunya virus; chronic viral infection; human immunology.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests D.W. is a consultant for Moderna. A.S. is a consultant for Alcimed, Arcturus, DarwinHealth, Desna Therapeutics, EmerVax, Gilead Sciences, Guggenheim Securities, Link University, and RiverVest Venture Partners. LJI has filed for patent protection for various aspects of T cell epitope and vaccine design work.
Figures




References
-
- Metz S.W., Pijlman G.P. In: Chikungunya Virus: Advances in Biology, Pathogenesis, and Treatment. Okeoma C.M., editor. Springer International Publishing; 2016. Function of Chikungunya Virus Structural Proteins; pp. 63–74. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

