CDX1 and CDX2 suppress colon cancer stemness by inhibiting β-catenin-facilitated formation of Pol II-DSIF-PAF1C complex
- PMID: 40399276
- PMCID: PMC12095478
- DOI: 10.1038/s41419-025-07737-3
CDX1 and CDX2 suppress colon cancer stemness by inhibiting β-catenin-facilitated formation of Pol II-DSIF-PAF1C complex
Abstract
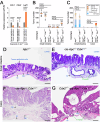
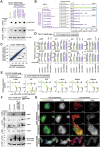
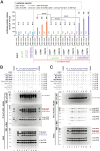
Homeobox transcription factors CDX1 and CDX2 (hereafter, CDX1/2) play key roles in determining the identity of intestinal epithelial cells and regulating their stem cell functions. However, the role of CDX1/2 in regulating colon cancer stemness and the underlying mechanisms are unclear. Here, we show that complete loss of Cdx1 or concurrent loss of Cdx1/2 increased the stemness and malignancy of intestinal tumors. Consistently, CDX1/2 reduced the expression of cancer stemness-related genes, including LGR5. CDX1/2 bound to the downstream region of the LGR5 transcription start site (TSS), a region where β-catenin also binds. Despite increased H3 acetylation and an open chromatin structure, CDX1/2 reduced the occupancy of DRB sensitivity-inducing factor (DSIF), RNA polymerase II-associated factor 1 (PAF1), and RNA polymerase II (Pol II) complexes around the LGR5 TSS. Through their homeodomains, CDX1/2 inhibited the β-catenin-facilitated formation of active Pol II complexes containing DSIF and PAF1 complexes by preventing the interaction between β-catenin and these complexes, in an additive manner. Our findings suggest that CDX1/2 cooperatively suppressed colonic tumorigenesis and cancer stemness by antagonizing β-catenin via the DSIF and PAF1 complexes. Additionally, DSIF and PAF1 complexes acted as transcriptional platforms that integrated and funneled both tumor-suppressive and oncogenic signals into the expression of genes that control colon cancer stemness.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethical approval: All methods in this study were carried out in accordance with relevant guidelines and regulations. All animal experiments in this study were approved by the Animal Care and Use Committee of FUKUI University (approval number R04023). This study did not involve human participants or patient-derived materials; therefore, ethical approvals and informed consents were not applicable.
Figures





References
-
- Mlodzik M, Gehring WJ. Expression of the caudal gene in the germ line of Drosophila: formation of an RNA and protein gradient during early embryogenesis. Cell. 1987;48:465–78. - PubMed
-
- Silberg DG, Swain GP, Suh ER, Traber PG. Cdx1 and cdx2 expression during intestinal development. Gastroenterology. 2000;119:961–71. - PubMed
-
- Kakizaki F, Aoki K, Miyoshi H, Carrasco N, Aoki M, Taketo MM. CDX transcription factors positively regulate expression of solute carrier family 5, member 8 in the colonic epithelium. Gastroenterology. 2010;138:627–35. - PubMed
-
- Chawengsaksophak K, James R, Hammond VE, Köntgen F, Beck F. Homeosis and intestinal tumours in Cdx2 mutant mice. Nature. 1997;386:84–87. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

