WorkflowHub: a registry for computational workflows
- PMID: 40399296
- PMCID: PMC12095652
- DOI: 10.1038/s41597-025-04786-3
WorkflowHub: a registry for computational workflows
Abstract
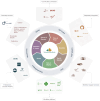
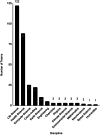
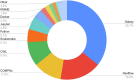
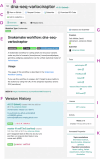
The rising popularity of computational workflows is driven by the need for repetitive and scalable data processing, sharing of processing know-how, and transparent methods. As both combined records of analysis and descriptions of processing steps, workflows should be reproducible, reusable, adaptable, and available. Workflow sharing presents opportunities to reduce unnecessary reinvention, promote reuse, increase access to best practice analyses for non-experts, and increase productivity. In reality, workflows are scattered and difficult to find, in part due to the diversity of available workflow engines and ecosystems, and because workflow sharing is not yet part of research practice. WorkflowHub provides a unified registry for all computational workflows that links to community repositories, and supports both the workflow lifecycle and making workflows findable, accessible, interoperable, and reusable (FAIR). By interoperating with diverse platforms, services, and external registries, WorkflowHub adds value by supporting workflow sharing, explicitly assigning credit, enhancing FAIRness, and promoting workflows as scholarly artefacts. The registry has a global reach, with hundreds of research organisations involved, and more than 800 workflows registered.
© 2025. UT-Battelle, LLC and the Authors.
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures

References
-
- Ferreira da Silva, R. et al. Workflows Community Summit 2022: A Roadmap Revolution 10.5281/zenodo.7750670 (2023).
-
- Amstutz, P., Mikheev, M., Crusoe, M. R., Tijanić, N. & Lampa, S. Existing Workflow systems. Common Workflow Language wiki. https://s.apache.org/existing-workflow-systems.
-
- Freudling, W. et al. Adaptive Data Reduction Workflows for Astronomy – The ESO Data Processing System (EDPS) 10.48550/ARXIV.2311.03822 (2023).
-
- McClure, J. E. et al. in Toward Real-Time Analysis of Synchrotron Micro-Tomography Data: Accelerating Experimental Workflows with AI and HPC (eds Nichols, J. et al.) Driving Scientific and Engineering Discoveries Through the Convergence of HPC, Big Data and AI, Vol. 1315 226–239 (Springer International Publishing, Cham, 2020). 10.1007/978-3-030-63393-6_15.
LinkOut - more resources
Full Text Sources
Research Materials

