Establishment of salivary tissue-organoid biorepository: characterizing salivary gland stem/progenitor cells and novel differentiation marker PSMA/FOLH1
- PMID: 40399315
- PMCID: PMC12095476
- DOI: 10.1038/s41536-025-00410-5
Establishment of salivary tissue-organoid biorepository: characterizing salivary gland stem/progenitor cells and novel differentiation marker PSMA/FOLH1
Abstract
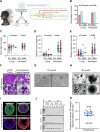
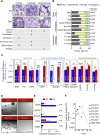
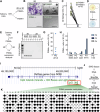
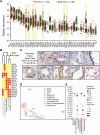
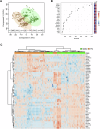
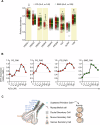
The salivary gland (SG) is vital for oral function and overall health through secretion of saliva. However salivary dysfunction due to aging, medications, autoimmune disorders, and cancer treatments poses significant challenges. We established the first diverse and clinically annotated salivary regenerative biobank at Mayo Clinic to study salivary gland stem/progenitor cells (SGSPCs). Optimization of cell isolation and progenitor assays revealed SGSPCs enriched within the CD24/EpCAM/CD49f+ and PSMA- phenotypes of both submandibular and parotid glands, with clonal differentiation assays highlighting heterogeneity. Induction of PSMA/FOLH1 expression was associated with SGSPC differentiation. Using mass spectrometry-based single cell proteomics, we identified 2461 proteins in SGSPC-enriched cells, including co-expressed cytokeratins, expressed in rare salivary ductal basal cells. Additionally, PRDX, a unique class of peroxiredoxin peroxidases enriched in SGSPCs, demonstrated H2O2-dependent growth, suggesting a role in salivary homeostasis. These findings provide a foundation for SGSPC research and potential regenerative therapies for salivary gland dysfunction.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


Similar articles
-
The Mayo Clinic Salivary Tissue-Organoid Biobanking: A Resource for Salivary Regeneration Research.bioRxiv [Preprint]. 2024 Feb 27:2024.02.23.581761. doi: 10.1101/2024.02.23.581761. bioRxiv. 2024. PMID: 38464033 Free PMC article. Preprint.
-
Comparative analysis reveals similarities between cultured submandibular salivary gland cells and liver progenitor cells.Springerplus. 2014 Apr 9;3:183. doi: 10.1186/2193-1801-3-183. eCollection 2014. Springerplus. 2014. PMID: 24790827 Free PMC article.
-
Salisphere derived c-Kit+ cell transplantation restores tissue homeostasis in irradiated salivary gland.Radiother Oncol. 2013 Sep;108(3):458-63. doi: 10.1016/j.radonc.2013.05.020. Epub 2013 Jun 13. Radiother Oncol. 2013. PMID: 23769181
-
Unveiling Stem Cell Heterogeneity Toward the Development of Salivary Gland Regenerative Strategies.Adv Exp Med Biol. 2019;1123:151-164. doi: 10.1007/978-3-030-11096-3_9. Adv Exp Med Biol. 2019. PMID: 31016599 Review.
-
Application of regenerative medicine to salivary gland hypofunction.Jpn Dent Sci Rev. 2021 Nov;57:54-59. doi: 10.1016/j.jdsr.2021.03.002. Epub 2021 Apr 27. Jpn Dent Sci Rev. 2021. PMID: 33995711 Free PMC article. Review.
References
-
- Pedersen, A. M. L., Sorensen, C. E., Proctor, G. B., Carpenter, G. H. & Ekstrom, J. Salivary secretion in health and disease. J. Oral. Rehabil.45, 730–746 (2018). - PubMed
-
- Mercadante, V., Al Hamad, A., Lodi, G., Porter, S. & Fedele, S. Interventions for the management of radiotherapy-induced xerostomia and hyposalivation: A systematic review and meta-analysis. Oral. Oncol.66, 64–74 (2017). - PubMed
-
- Taïeb, D. et al. PSMA-Targeted Radionuclide Therapy and Salivary Gland Toxicity: Why Does It Matter?. J. Nucl. Med.59, 747–748 (2018). - PubMed
-
- Emperumal, C. P. et al. Oral Toxicities of PSMA-Targeted Immunotherapies for The Management of Prostate Cancer. Clin. Genitourinary Cancer22, 380–384 (2023). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

