Small-molecule screen in C. elegans identifies benzenesulfonamides as inhibitors of microsporidia spores
- PMID: 40399462
- PMCID: PMC12095817
- DOI: 10.1038/s44259-025-00116-0
Small-molecule screen in C. elegans identifies benzenesulfonamides as inhibitors of microsporidia spores
Abstract
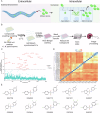
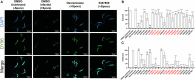
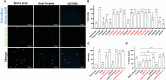
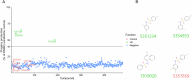
Microsporidia, a large group of fungal-related intracellular parasites, infect several economically significant animals, leading to substantial economic losses. As currently available anti-microsporidia therapies are either ineffective or come with numerous adverse effects, there is a need for alternative microsporidia inhibitors. Here we screen a subset of the ChemBridge DIVERset library, comprising 2500 diverse compounds, using Caenorhabditis elegans infected with its natural microsporidian parasite, Nematocida parisii. By testing these compounds at 60 μM in 96-well assay plates, we identified 26 hits that restored the ability of C. elegans to produce progeny in the presence of N. parisii. We confirmed that out of 20 tested compounds, 18 ChemBridge compounds effectively inhibit N. parisii infection in C. elegans. Of these 18, 10 were benzenesulfonamide derivatives which inhibit microsporidia infection by inactivating spores. We screened an additional 475-compound benzenesulfonamide library, successfully identifying three compounds that are effective at a lower concentration than the initial hits. We further show that one benzenesulfonamide compound displays inhibitory activity against several species of microsporidia, inhibiting infection of species belonging to the Nematocida, Enterocytozoon, and Encephalitozoon genera. Together our results suggest that benzenesulfonamides are a potential scaffold for the development of microsporidia antiseptics.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




Similar articles
-
Screening of the Pandemic Response Box identifies anti-microsporidia compounds.PLoS Negl Trop Dis. 2023 Dec 8;17(12):e0011806. doi: 10.1371/journal.pntd.0011806. eCollection 2023 Dec. PLoS Negl Trop Dis. 2023. PMID: 38064503 Free PMC article.
-
High-throughput small molecule screen identifies inhibitors of microsporidia invasion and proliferation in C. elegans.Nat Commun. 2022 Sep 26;13(1):5653. doi: 10.1038/s41467-022-33400-y. Nat Commun. 2022. PMID: 36163337 Free PMC article.
-
A Large Collection of Novel Nematode-Infecting Microsporidia and Their Diverse Interactions with Caenorhabditis elegans and Other Related Nematodes.PLoS Pathog. 2016 Dec 12;12(12):e1006093. doi: 10.1371/journal.ppat.1006093. eCollection 2016 Dec. PLoS Pathog. 2016. PMID: 27942022 Free PMC article.
-
Host-Microsporidia Interactions in Caenorhabditis elegans, a Model Nematode Host.Microbiol Spectr. 2016 Oct;4(5). doi: 10.1128/microbiolspec.FUNK-0003-2016. Microbiol Spectr. 2016. PMID: 27763260 Review.
-
Insights from C. elegans into Microsporidia Biology and Host-Pathogen Relationships.Exp Suppl. 2022;114:115-136. doi: 10.1007/978-3-030-93306-7_5. Exp Suppl. 2022. PMID: 35544001 Free PMC article. Review.
Cited by
-
Identification of natural products and synthetic analogs which inhibit microsporidia spores and prevent infection.bioRxiv [Preprint]. 2025 Aug 6:2025.08.06.669004. doi: 10.1101/2025.08.06.669004. bioRxiv. 2025. PMID: 40799553 Free PMC article. Preprint.
References
-
- Keeling, P. J. & Fast, N. M. Microsporidia: biology and evolution of highly reduced intracellular parasites. Annu. Rev. Microbiol.56, 93–116 (2002). - PubMed
-
- Willis, A. R. & Reinke, A. W. Factors that determine microsporidia infection and host specificity. Exp. Suppl. 2012114, 91–114 (2022). - PubMed
-
- Vossbrinck, C. R., Andreadis, T. G. & Weiss, L. M. Phylogenetics: taxonomy and the microsporidia as derived fungi. in Opportunistic Infections: Toxoplasma, Sarcocystis, and Microsporidia (eds. Lindsay, D. S. & Weiss, L. M.) vol. 9 189–213 (Springer, 2004).
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

