Genomic landscape of multiple myeloma and its precursor conditions
- PMID: 40399554
- PMCID: PMC12165861
- DOI: 10.1038/s41588-025-02196-0
Genomic landscape of multiple myeloma and its precursor conditions
Abstract
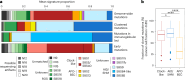
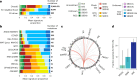
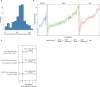
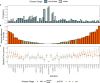
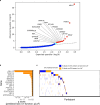
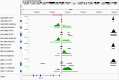
Reliable strategies to capture patients at risk of progression from precursor stages of multiple myeloma (MM) to overt disease are still missing. We assembled a comprehensive collection of MM genomic data comprising 1,030 patients (218 with precursor conditions) that we used to identify recurrent coding and non-coding candidate drivers as well as significant hotspots of structural variation. We used those drivers to define and validate a simple 'MM-like' score, which we could use to place patients' tumors on a gradual axis of progression toward active disease. Our MM precursor genomic map provides insights into the time of initiation and cell-of-origin of the disease, order of acquisition of genomic alterations and mutational processes found across the stages of transformation. Taken together, we highlight here the potential of genome sequencing to better inform risk assessment and monitoring of MM precursor conditions.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: J.-B.A., A.K.D., G.G. and I.M.G. are inventors on patent applications for MinimuMM-seq and for the MM-like score. A.K.D. and E.D.L. have consulted for and received honoraria from Menarini Silicon Biosystems. G.G. receives research funds from IBM, Pharmacyclics/Abbvie, Bayer, Genentech, Calico, Ultima Genomics, Inocras, Google, Kite and Novartis, and is also an inventor on patent applications filed by the Broad Institute related to MSMuTect, MSMutSig, POLYSOLVER, SignatureAnalyzer-GPU, MSEye, MinimuMM-seq, and DLBclass. He is a founder, consultant and holds private equity in Scorpion Therapeutics; he is also a founder of and holds privately held equity in Predicta Biosciences. He was also a consultant to Merck. I.M.G. has consulted for Bristol-Myers Squibb, AstraZeneca, Amgen, Curio Science, Sanofi, Janssen, Pfizer, Menarini Silicone Biosystems, Aptitude Health, GlaxoSmithKline, AbbVie, Adaptive Biotechnologies, Window Therapeutics and Regeneron. She has received honoraria or speaker fees from Vor Biopharma, Janssen, MJH Life Sciences, Novartis, Takeda, Amgen, Regeneron, Curio Science, Standard Biotools and Physicians’ Education Resource. She is a founder and executive board member of and holds private equity in Predicta Biosciences. Her spouse is the chief medical officer of and holds private equity in Disc Medicine. R.S.P. is a founder, consultant and holds equity in Predicta Biosciences. E.Z. has received honoraria and served in advisory roles for Janssen, Bristol-Myers Squibb, Sanofi, Amgen, GlaxoSmithKline, Pfizer, Oncopeptides and Menarini–Stemline. The other authors declare no competing interests.
Figures









References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

