Genome-wide association study of long COVID
- PMID: 40399555
- PMCID: PMC12165857
- DOI: 10.1038/s41588-025-02100-w
Genome-wide association study of long COVID
Abstract
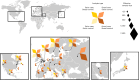
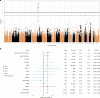
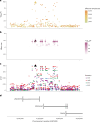
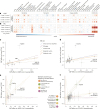
Infections can lead to persistent symptoms and diseases such as shingles after varicella zoster or rheumatic fever after streptococcal infections. Similarly, severe acute respiratory syndrome coronavirus 2 (SARS‑CoV‑2) infection can result in long coronavirus disease (COVID), typically manifesting as fatigue, pulmonary symptoms and cognitive dysfunction. The biological mechanisms behind long COVID remain unclear. We performed a genome-wide association study for long COVID including up to 6,450 long COVID cases and 1,093,995 population controls from 24 studies across 16 countries. We discovered an association of FOXP4 with long COVID, independent of its previously identified association with severe COVID-19. The signal was replicated in 9,500 long COVID cases and 798,835 population controls. Given the transcription factor FOXP4's role in lung physiology and pathology, our findings highlight the importance of lung function in the pathophysiology of long COVID.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: S.B. has ownerships in Intomics A/S, Hoba Therapeutics Aps, Novo Nordisk A/S, Lundbeck A/S, ALK abello A/S, Eli Lilly and Co and is managing board memberships in Proscion A/S and Intomics A/S. A.B., K.M.S.B., S.W., N.L.W., F.T., E.S. and E.T.C. are employees of Helix. A.D. received an honorarium from Gilead Sciences. A.L.G. and C.J. have funded research collaborations with Orion for collaborative research projects outside the submitted work. T.H. and H.E.O.B. have options in Sano Genetics. P.J.S. is a shareholder of Sano Genetics. T.H.K. has received consulting fees from Albireo, Boehringer Ingelheim, MSD and Falk Pharma. K.U.L. is cofounder and member of the scientific board of LAMPseq Diagnostics GmbH. T.N. has received speaking fee from Boehringer Ingelheim for talks unrelated to this research. M.E.K.N. is a current employee of Novartis Pharma AG. J.B.R.’s institution has received investigator-initiated grant funding from Eli Lilly, GlaxoSmithKline and Biogen for projects unrelated to this research. He is the CEO of 5 Prime Sciences ( www.5primesciences.com ), which provides research services for biotech, pharma and venture capital companies for projects unrelated to this research. V.F. is an employee of 5 Prime Sciences. C.D.S. reports grants and personal fees from AstraZeneca, Janssen-Cilag and ViiV Healthcare, personal fees and nonfinancial support from BBraun Melsungen, grants, personal fees and nonfinancial support from Gilead Sciences, personal fees from BioNtech, Eli Lilly, Formycon, Pfizer, Roche, Apeiron, GSK, Molecular partners, SOBI, AbbVie, MSD and Synairgen and grants from Cepheid. L.V.W. reports research funding from GlaxoSmithKline, Genentech and Orion Pharma, and consultancy for Galapagos and GlaxoSmithKline, outside of the submitted work. J.W. is a consultant for Roboscreen GmbH, Biogen GmbH, Immungenetics AG, Noselab GmbH, Roche Diagnostics International, Roche Pharma AG, Janssen-Cilag GmbH, Eisai GmbH, Boehringer Ingelheim and Lilly Deutschland GmbH and has received honoraries from Eisai GmbH, Biogen GmbH, AGNP e. V., Veranex, Med Update GmbH, Guangzhou Gloryren Medical Technology (China), Pfizer Pharma GmbH, Fachverband Rheumatologische Fachassistenz e. V., AWO Psychiatrie Akademie gGmbH, Neuroakademie E. V., Beijing Yibai Science und Technology Ltd., Abbott Laboratories GmbH, Lilly Deutschland GmbH, Simon & Kucher and streamedup! GmbH. The other authors declare no competing interests.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

