Multigenerational cell tracking of DNA replication and heritable DNA damage
- PMID: 40399682
- PMCID: PMC12176655
- DOI: 10.1038/s41586-025-08986-0
Multigenerational cell tracking of DNA replication and heritable DNA damage
Abstract
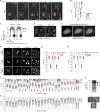
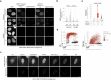
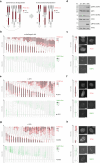
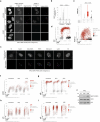
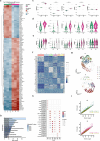
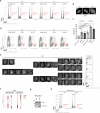
Cell heterogeneity is a universal feature of life. Although biological processes affected by cell-to-cell variation are manifold, from developmental plasticity to tumour heterogeneity and differential drug responses, the sources of cell heterogeneity remain largely unclear1,2. Mutational and epigenetic signatures from cancer (epi)genomics are powerful for deducing processes that shaped cancer genome evolution3-5. However, retrospective analyses face difficulties in resolving how cellular heterogeneity emerges and is propagated to subsequent cell generations. Here, we used multigenerational single-cell tracking based on endogenously labelled proteins and custom-designed computational tools to elucidate how oncogenic perturbations induce sister cell asymmetry and phenotypic heterogeneity. Dual CRISPR-based genome editing enabled simultaneous tracking of DNA replication patterns and heritable endogenous DNA lesions. Cell lineage trees of up to four generations were tracked in asynchronously growing cells, and time-resolved lineage analyses were combined with end-point measurements of cell cycle and DNA damage markers through iterative staining. Besides revealing replication and repair dynamics, damage inheritance and emergence of sister cell heterogeneity across multiple cell generations, through combination with single-cell transcriptomics, we delineate how common oncogenic events trigger multiple routes towards polyploidization with distinct outcomes for genome integrity. Our study provides a framework to dissect phenotypic plasticity at the single-cell level and sheds light onto cellular processes that may resemble early events during cancer development.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures











References
-
- Koh, G., Degasperi, A., Zou, X., Momen, S. & Nik-Zainal, S. Mutational signatures: emerging concepts, caveats and clinical applications. Nat. Rev. Cancer21, 619–637 (2021). - PubMed
-
- Ushijima, T., Clark, S. J. & Tan, P. Mapping genomic and epigenomic evolution in cancer ecosystems. Science373, 1474–1479 (2021). - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

