Transcriptomic analysis of DENV-2-infected human dermal fibroblasts identified potential mechanisms that suppressed ZIKV replication during sequential coinfection
- PMID: 40399994
- PMCID: PMC12096689
- DOI: 10.1186/s12985-025-02769-9
Transcriptomic analysis of DENV-2-infected human dermal fibroblasts identified potential mechanisms that suppressed ZIKV replication during sequential coinfection
Abstract
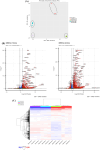
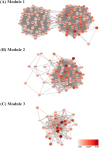
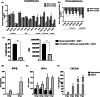
Dengue virus (DENV) and Zika virus (ZIKV) are closely related flaviviruses which are transmitted by the same species of mosquitoes. Due to overlapping geographic distributions and transmission vectors, cases of DENV-ZIKV coinfection have been reported. However, the impact of coinfection on disease outcomes remains unclear. In this study, an in vitro model of DENV-ZIKV coinfection was developed using the primary human dermal fibroblasts (HDFs). The interaction between DENV-2 and ZIKV during sequential coinfection revealed that prior DENV-2 infection significantly suppressed ZIKV RNA accumulation in the culture supernatant. Transcriptomic profile in response to DENV-2 infection suggested three hypothetical pathways that potentially interfere with ZIKV replication. The first mechanism is prior DENV infection drove HDFs into an antiviral state through upregulation of genes involving innate immune response pathways, including PRR signaling, type I and type II IFN signaling, ISG activity, and cytokine/chemokine activity. This state significantly enhanced resistance to subsequent ZIKV infection in both infected cells and uninfected neighboring cells. The second potential pathway is inhibition of viral entry. This was supported by DENV-2-infected HDFs significantly suppressed expression of ZIKV receptor and reduced expression of genes involving in clathrin-mediated endocytosis. This can interfere with entry of ZIKV into host cells. The last possible mechanism is driving cells into cell cycle arrest, as DENV-2 infection downregulated genes related to cell cycle progression, which may hinder ZIKV replication. These findings partly unfold the interplay between DENV and ZIKV at the entry site which may explain the disease outcome of DENV-ZIKV coinfection.
Keywords: Coinfection; Dengue virus; Human dermal fibroblasts; Superinfection exclusion; Transcriptomic profiling; Zika virus.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

