Role of LEAFLESS, an AP2/ERF family transcription factor, in the regulation of trichome specialized metabolism
- PMID: 40400206
- PMCID: PMC12177301
- DOI: 10.1111/nph.70198
Role of LEAFLESS, an AP2/ERF family transcription factor, in the regulation of trichome specialized metabolism
Abstract
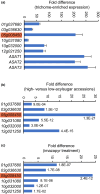
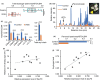
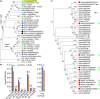
Acylsugars, specialized metabolites produced by trichomes of many solanaceous species, provide protection against biotic and abiotic stresses. Many acylsugar metabolic enzymes have been identified; however, regulatory factors remain unknown. Our multidisciplinary approaches identified LEAFLESS (APETALA 2/ ETHYLENE RESPONSE FACTOR (AP2/ERF) family member) as a positive regulator of acylsugar biosynthesis. Virus-induced gene silencing (VIGS) of LEAFLESS in Solanum pennellii (SpLFS/Sopen05g008450) revealed its distinct roles in two related but separate processes: acylsugar biosynthesis and trichome development. Most acylsugar (and several flavonoid) metabolic genes were downregulated in SpLFS-silenced plants and showed strong co-expression with SpLFS. Phylogenetic and additional data analyses indicated trichome-enriched expression of SpLFS orthologs in other acylsugar-producing solanaceous species, and VIGS of SpLFS orthologs in Nicotiana benthamiana reduced acylsugar production. Transcriptional reporter showed expression of SpLFS in type I/IV trichome tip cells, the site of acylsugar biosynthesis. Electrophoretic mobility shift assays indicated that SpLFS directly binds to promoters of several acylsugar (and flavonoid) metabolic genes. Additionally, data mining suggested remarkable spatiotemporal functional diversity: from coordinating leaf initiation at incipient primordia (previously reported for the S. lycopersicum ortholog SlLFS/Solyc05g013540) to regulating trichome specialized metabolism (acylsugar and flavonoid). Our work highlights a critical role of LEAFLESS in trichome specialized metabolism, paving the way to disentangle the acylsugar regulatory network.
Keywords: AP2/ERF; LEAFLESS; Solanaceae; WGCNA; acylsugar; flavonoid; trichome; virus‐induced gene silencing.
© 2025 The Author(s). New Phytologist © 2025 New Phytologist Foundation.
Conflict of interest statement
None declared.
Figures



Similar articles
-
Glandless, a tomato HD-ZIP transcription factor, is important for the gland formation of type VI trichomes.Plant J. 2025 Jul;123(1):e70308. doi: 10.1111/tpj.70308. Plant J. 2025. PMID: 40605474 Free PMC article.
-
Tomato root specialized metabolites evolved through gene duplication and regulatory divergence within a biosynthetic gene cluster.Sci Adv. 2024 Apr 26;10(17):eadn3991. doi: 10.1126/sciadv.adn3991. Epub 2024 Apr 24. Sci Adv. 2024. PMID: 38657073 Free PMC article.
-
Specialized metabolism by trichome-enriched Rubisco and fatty acid synthase components.Plant Physiol. 2023 Feb 12;191(2):1199-1213. doi: 10.1093/plphys/kiac487. Plant Physiol. 2023. PMID: 36264116 Free PMC article.
-
Tip of the trichome: evolution of acylsugar metabolic diversity in Solanaceae.Curr Opin Plant Biol. 2019 Jun;49:8-16. doi: 10.1016/j.pbi.2019.03.005. Epub 2019 Apr 19. Curr Opin Plant Biol. 2019. PMID: 31009840 Free PMC article. Review.
-
Deciphering the ″Developmental Code″: A Comprehensive Review of Regulatory Mechanisms in Tomato Trichomes.J Agric Food Chem. 2025 Jul 16;73(28):17358-17373. doi: 10.1021/acs.jafc.5c02904. Epub 2025 Jul 1. J Agric Food Chem. 2025. PMID: 40591814 Review.
References
-
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B: Statistical Methodology 57: 289–300.
-
- Blauth SL, Churchill GA, Mutschler MA. 1998. Identification of quantitative trait loci associated with acylsugar accumulation using intraspecific populations of the wild tomato, Lycopersicon pennellii. Theoretical and Applied Genetics 96: 458–467. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

