3-D substructure search by transitive closure in AlphaFold database
- PMID: 40400345
- PMCID: PMC12095923
- DOI: 10.1002/pro.70169
3-D substructure search by transitive closure in AlphaFold database
Abstract
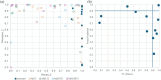
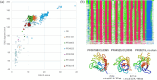
Identifying structural relationships between proteins is crucial for understanding their functions and evolutionary histories. We present ISS_ProtSci, a Python package designed for structural similarity searches within the AlphaFold Database v2 (AFDB2). ISS_ProtSci incorporates DaliLite to identify geometrically similar structures and uses a transitive closure algorithm to iteratively explore neighboring shells of proteins. The precomputed all-against-all comparisons generated by Foldseek, chosen for its speed, are validated by DaliLite for precision. Search results are annotated with metadata from UniProtKB and Pfam protein family classifications, using hmmsearch to identify protein domains. Outputs, including Dali pairwise alignment data, are provided in TSV format for easy filtering and analysis. Our method offers a significant improvement in recall over existing tools like Foldseek, especially in detecting more distantly related proteins. This is particularly valuable in structurally diverse protein families where traditional sequence-based or fast structural methods struggle. ISS_ProtSci delivers practical runtimes and flexibility, allowing users to input a PDB file, define the minimum size of the common core, and evaluate results using Pfam clans. In evaluating our method across 12 test cases based on Pfam clans, we achieved over 99% recall of relevant proteins, even in challenging cases where Foldseek's recall dropped below 50%. ISS_ProtSci not only identifies closely related proteins but also uncovers previously unrecognized structural relationships, contributing to more accurate protein family classifications. The software can be downloaded from http://ekhidna2.biocenter.helsinki.fi/ISS_ProtSci/.
Keywords: Dali; Foldseek; Pfam; protein space; superfamily.
© 2025 The Author(s). Protein Science published by Wiley Periodicals LLC on behalf of The Protein Society.
Figures





Similar articles
-
Benchmarking fold detection by DaliLite v.5.Bioinformatics. 2019 Dec 15;35(24):5326-5327. doi: 10.1093/bioinformatics/btz536. Bioinformatics. 2019. PMID: 31263867
-
DALI shines a light on remote homologs: One hundred discoveries.Protein Sci. 2023 Jan;32(1):e4519. doi: 10.1002/pro.4519. Protein Sci. 2023. PMID: 36419248 Free PMC article.
-
Searching protein structure databases with DaliLite v.3.Bioinformatics. 2008 Dec 1;24(23):2780-1. doi: 10.1093/bioinformatics/btn507. Epub 2008 Sep 25. Bioinformatics. 2008. PMID: 18818215 Free PMC article.
-
Dali server: structural unification of protein families.Nucleic Acids Res. 2022 Jul 5;50(W1):W210-W215. doi: 10.1093/nar/gkac387. Nucleic Acids Res. 2022. PMID: 35610055 Free PMC article.
-
Comparison of proteins based on segments structural similarity.Acta Biochim Pol. 2004;51(1):161-72. Acta Biochim Pol. 2004. PMID: 15094837 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

