Complete chloroplast genome sequence of Quercus salicina Blume (Fagaceae)
- PMID: 40400641
- PMCID: PMC12093796
- DOI: 10.1080/23802359.2025.2503396
Complete chloroplast genome sequence of Quercus salicina Blume (Fagaceae)
Abstract
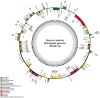
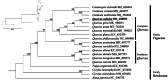
Quercus salicina Blume 1851, an evergreen broad-leaved species belonging to the family Fagaceae, is primarily distributed from southern Korea to Japan. In this study, we comprehensively analyzed the chloroplast genome of Q. salicina. Its genome, with a quadripartite structure, spanned 160,801 bp and had a GC content of 36.58%. Annotation revealed 131 genes, including 86 protein-coding genes, eight rRNA genes, and 37 tRNA genes. These intricacies in the genome were further confirmed by the presence of introns in multiple genes. Phylogenetic analysis of 19 Fagales species elucidated their evolutionary relationships, with Fagus engleriana branching early and Q. salicina showing a close relationship with Q. gilva. This study enhances our understanding of the genetic makeup of Q. salicina and its evolutionary dynamics within the Fagales.
Keywords: Chloroplast genome; Fagaceae; Phylogenetic analysis; Quercus salicina.
© 2025 National Institute of Forest Science. Published by Informa UK Limited, trading as Taylor & Francis Group.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures

References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
