Defining lung adenocarcinoma subtypes with glucocorticoid-related genes and constructing a prognostic index for immunotherapy guidance
- PMID: 40400930
- PMCID: PMC12090106
- DOI: 10.21037/jtd-24-1083
Defining lung adenocarcinoma subtypes with glucocorticoid-related genes and constructing a prognostic index for immunotherapy guidance
Erratum in
-
Erratum: Defining lung adenocarcinoma subtypes with glucocorticoid-related genes and constructing a prognostic index for immunotherapy guidance.J Thorac Dis. 2025 Nov 30;17(11):10588. doi: 10.21037/jtd-2025b-09. Epub 2025 Nov 26. J Thorac Dis. 2025. PMID: 41376996 Free PMC article.
Abstract
Background: Several studies have shown that glucocorticoid-related genes (GCGs) play a crucial role in cancer. However, the mechanism of GCGs in lung adenocarcinoma (LUAD) is not fully understood. This study aimed to identify distinct subtypes of LUAD by integrating GCGs and to develop prognostic models for precise prognosis prediction and immunotherapy guidance.
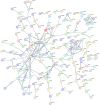
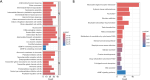
Methods: In this study, sample data of LUAD were collected from The Cancer Genome Atlas (TCGA) database, and unsupervised clustering was used to identify LUAD subtypes with different GCGs characteristics. Survival-related genes were screened by differential expression analysis and protein-protein interaction (PPI) network analysis. After that, the least absolute shrinkage and selection operator (LASSO) combined with Cox regression analysis was used to establish the prognosis model. Differences in the immune microenvironment of different risk groups were analyzed, and Tumor Immune Dysfunction and Exclusion (TIDE) was used to predict the response of patients to immunotherapy. Finally, the CellMiner database was used to predict potential drugs.
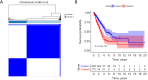
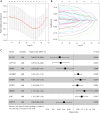
Results: Two subtypes of LUAD were identified, namely cluster 1 (high survival rate) and cluster 2 (low survival rate). A prognostic model was constructed based on 9 characteristic genes, including CLCA1, CYP17A1, GRIA2, IGFBP1, IGF2BP1, NTSR1, RPE65, VGF, and WNT16, and the prognosis of LUAD patients was positively predicted. There were differences in the immune microenvironment of different risk LUAD patients, and high-risk LUAD patients may benefit less from immunotherapy. BGB-283 was a candidate for LUAD targeting VGF.
Conclusions: Our study elucidates the impact of GCGs on LUAD prognosis and immune responses, offering insights for prognostic forecasting and immunotherapeutic strategies for LUAD patients.
Keywords: Glucocorticoid-related genes (GCGs); immunotherapy; lung adenocarcinoma (LUAD); prognostic models; subtypes.
Copyright © 2025 AME Publishing Company. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-24-1083/coif). The authors have no conflicts of interest to declare.
Figures




References
LinkOut - more resources
Full Text Sources
