Construction of a multigenic diagnostic, prognostic, and immune infiltration model with methylation-associated regulators in esophageal squamous cell carcinoma
- PMID: 40400969
- PMCID: PMC12090105
- DOI: 10.21037/jtd-2025-341
Construction of a multigenic diagnostic, prognostic, and immune infiltration model with methylation-associated regulators in esophageal squamous cell carcinoma
Abstract
Background: Methylation-related regulators may be involved in the prognostic prediction of esophageal squamous cell carcinoma (ESCC). Our study aimed to apply bioinformatics to screen methylation-related regulators for the construction of a prognostic model for patients with ESCC and to assess their diagnostic value and correlation with immune infiltration.
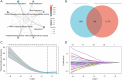
Methods: Prognosis-related genes were identified from The Cancer Genome Atlas (TCGA) database. Methylation-associated genes were filtered using the GeneCards database. We used the least absolute shrinkage and selection operator (LASSO) and Cox proportional hazards regression to identify the prognostic indicators. We applied the single-sample gene set enrichment analysis (ssGSEA) to clarify the relationship between prognostic indicators and immune infiltration in patients with ESCC (n=82).
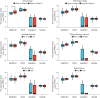
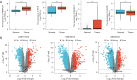
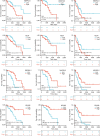
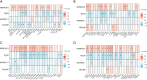
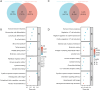
Results: We constructed a prognostic model using methylation-related regulators homocysteine-inducible ER protein with ubiquitin-like domain 1 (HERPUD1), trans-2,3-enoyl-CoA reductase (TECR), melanoma antigen gene A11 (MAGEA11), and NOP2/Sun RNA methyltransferase 6 (NSUN6) to evaluate the prognosis of patients with ESCC. A higher prognostic risk score was associated with shorter overall survival (OS) in patients with ESCC [hazard ratio (HR) =5.77, 95% confidence interval (CI): 2.13-15.58; P<0.001]. Time-dependent area under the curve (AUC) analysis revealed that HERPUD1, TECR, MAGEA11, and NSUN6 had high prognostic predictive value at different time points. Furthermore, we found that the combined diagnostic model based on HERPUD1, TECR, MAGEA11, and NSUN6 had excellent diagnostic efficacy for ESCC (AUC =0.911; 95% CI: 0.888-0.935). Finally, the ssGSEA algorithm showed that HERPUD1 was significantly positively correlated with immune infiltration at both the cellular and genetic levels, while TECR showed a significant negative correlation with immune infiltration levels.
Conclusions: Our prognostic model, built with the methylation-related regulators HERPUD1, TECR, MAGEA11, and NSUN6, could effectively predict prognosis in patients with ESCC, enhance diagnostic efficacy, and reflect immune cell infiltration in their microenvironment. Our findings are hypothesis generating and larger confirmatory studies are needed to validate our results.
Keywords: Esophageal squamous cell carcinoma (ESCC); diagnosis; immune infiltration; methylation; prognosis.
Copyright © 2025 AME Publishing Company. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-2025-341/coif). S.M. reports grants from Ipsen biopharmaceuticals; consulting fees from Merck, Eisai, BeiGene, BMS; payment or honoraria from Integrity CE, LLC, and Academy for Continued Healthcare Learning; outside the submitted work; and serves as board member of Esophageal Cancer Action Network and panel member of NCCN. A.S. reports consulting or advisory board role with AstraZeneca, Bristol-Myers Squibb, Merck, Exelixis, Pfizer, Xilio therapeutics, Taiho, Amgen, Autem therapeutics, KAHR medical, Arcus therapeutics, Regeneron, Replimune and Daiichi Sankyo; institutional research funding from AstraZeneca, Bristol-Myers Squibb, Merck, Clovis, Exelixis, Actuate therapeutics, Incyte Corporation, Daiichi Sankyo, Five prime therapeutics, Amgen, Innovent biologics, Dragonfly therapeutics, Oxford Biotherapeutics, Replimune, Phanes therapeutics, Arcus therapeutics, Regeneron and KAHR medical, outside the submitted work. The other authors have no conflicts of interest to declare.
Figures



Similar articles
-
Comprehensive Analysis of PD-L1 Expression, Immune Infiltrates, and m6A RNA Methylation Regulators in Esophageal Squamous Cell Carcinoma.Front Immunol. 2021 May 12;12:669750. doi: 10.3389/fimmu.2021.669750. eCollection 2021. Front Immunol. 2021. PMID: 34054840 Free PMC article.
-
Estimating the prognosis of esophageal squamous cell carcinoma based on The Cancer Genome Atlas (TCGA) of m6A methylation-associated genes.J Gastrointest Oncol. 2022 Feb;13(1):1-12. doi: 10.21037/jgo-21-686. J Gastrointest Oncol. 2022. PMID: 35284129 Free PMC article.
-
A novel disulfidptosis-related LncRNA prognostic risk model: predicts the prognosis, tumor microenvironment and drug sensitivity in esophageal squamous cell carcinoma.BMC Gastroenterol. 2024 Nov 27;24(1):437. doi: 10.1186/s12876-024-03530-2. BMC Gastroenterol. 2024. PMID: 39604874 Free PMC article.
-
A novel immune-related gene signature predicts survival in esophageal squamous cell carcinoma.Transl Cancer Res. 2021 May;10(5):2354-2367. doi: 10.21037/tcr-20-2665. Transl Cancer Res. 2021. PMID: 35116551 Free PMC article.
-
Development and Validation of a Prognostic Gene Signature Correlated With M2 Macrophage Infiltration in Esophageal Squamous Cell Carcinoma.Front Oncol. 2021 Dec 3;11:769727. doi: 10.3389/fonc.2021.769727. eCollection 2021. Front Oncol. 2021. PMID: 34926275 Free PMC article.
References
LinkOut - more resources
Full Text Sources
