Transcription regulation by RNA-induced structural strain in duplex DNA
- PMID: 40401557
- PMCID: PMC12096072
- DOI: 10.1093/nar/gkaf429
Transcription regulation by RNA-induced structural strain in duplex DNA
Abstract
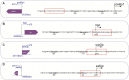
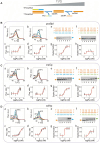
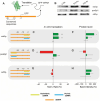
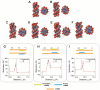
Non-coding RNAs belong to a heterogenous family that, among other functions, acts as a biomolecular regulator of gene expression. In particular, lncRNAs, which are estimated to be as numerous as coding RNAs in humans, are thought to interact with genomic DNA to form triple helices. However, experimental evidence of their involvement with processes, such as chromatin structure dynamics or RNA transcription, is still missing. Here, a mechanism of transcription enhancement/inhibition is described, where hybrid RNA-DNA triplexes regulate transcription rates in Escherichia coli promoter-based designed architectures. Sequences associated with triplexes were identified in a library of bacterial promoters and characterized in vitro, followed by a synthetic biology approach to verify their ability to control transcription and translation. A model of the triplex-promoter complex was produced showing that transcription enhancement is due to a distortion of the duplex DNA as a consequence of its conjugation with RNA in the triplex assembly. These results point at a mechanism of RNA function that is still unknown and could be common in more complex organisms, such as metazoans including mammals, where non-coding RNAs are more abundant and are believed to play a fundamental role in determining hetero/euchromatin and transcription modulation.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Conflict of interest statement
The authors declare no competing interest.
Figures


Similar articles
-
Genome-wide computational analysis of potential long noncoding RNA mediated DNA:DNA:RNA triplexes in the human genome.J Transl Med. 2017 Sep 2;15(1):186. doi: 10.1186/s12967-017-1282-9. J Transl Med. 2017. PMID: 28865451 Free PMC article.
-
Detection of RNA-DNA binding sites in long noncoding RNAs.Nucleic Acids Res. 2019 Apr 8;47(6):e32. doi: 10.1093/nar/gkz037. Nucleic Acids Res. 2019. PMID: 30698727 Free PMC article.
-
Practical Guidance in Genome-Wide RNA:DNA Triple Helix Prediction.Int J Mol Sci. 2020 Jan 28;21(3):830. doi: 10.3390/ijms21030830. Int J Mol Sci. 2020. PMID: 32012884 Free PMC article.
-
RNA-DNA Triplex Formation by Long Noncoding RNAs.Cell Chem Biol. 2016 Nov 17;23(11):1325-1333. doi: 10.1016/j.chembiol.2016.09.011. Epub 2016 Oct 20. Cell Chem Biol. 2016. PMID: 27773629 Review.
-
Biophysical Investigation of RNA ⋅ DNA : DNA Triple Helix and RNA : DNA Heteroduplex Formation by the lncRNAs MEG3 and Fendrr.Chembiochem. 2024 May 17;25(10):e202400049. doi: 10.1002/cbic.202400049. Epub 2024 Apr 29. Chembiochem. 2024. PMID: 38456652 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

