Transcriptomic insights into Candida albicans adaptation to an anaerobic environment
- PMID: 40401963
- PMCID: PMC12210919
- DOI: 10.1128/spectrum.03024-24
Transcriptomic insights into Candida albicans adaptation to an anaerobic environment
Abstract
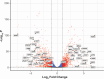
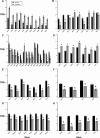
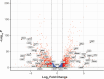
Candida albicans is a clinically significant fungal pathogen capable of adapting to diverse host environments, including steep oxygen gradients ranging from ~21% oxygen to anaerobic. The ability to withstand varied oxygen levels is paramount to establishing colonization and persisting in host niches, and oxygen deprivation can also augment antifungal resistance. In this study, we used RNA sequencing to compare the global transcriptomic profiles of two strains of C. albicans (SC5314 and CHN1) grown purely anaerobically to those grown aerobically. In C. albicans SC5314, we observed a strong induction of the alternative oxidase AOX2 and several genes encoding subunits of mitochondrial enzyme complexes I, II, and V, signifying a shift to alternative respiration. Consistent with the diminished ATP production from this process, there was a significant downregulation of genes associated with growth and metabolism, including histones and ribosomal proteins, as well as chitinases and other genes involved in cell wall remodeling. Interestingly, the anaerobic C. albicans cultures had decreased expression of candidalysin (ECE1) and other virulence factors, contrasting with other studies reporting enhanced pathogenicity under oxygen deprivation. There were a greater number of significantly upregulated genes in C. albicans CHN1 compared to SC5314; however, most of the top 50 upregulated genes under anaerobic conditions were consistent between the two strains. The predominant difference in down-regulated genes between the two strains could be mapped to differences in hyphal transformation under aerobic conditions. Overall, our study provides a window into the molecular mechanisms of C. albicans adaptation between aerobic to anaerobic environments.IMPORTANCECandida albicans is a leading cause of fungal infections in humans, posing significant clinical challenges due to its remarkable adaptability and increasing antifungal resistance. Anaerobic environments can promote antifungal resistance, necessitating a deeper understanding of how C. albicans adapts to anoxia. While much research has been done to identify mechanisms underlying adaptation to hypoxia (i.e., low oxygen), this is the first study evaluating the global transcriptomic response of C. albicans to anoxia (no oxygen). Here, we uncover key transcriptomic changes that enable C. albicans to survive in the absence of oxygen, which are distinct from those identified under hypoxic conditions. Our research addresses a gap in current knowledge that may be exploited for combatting antifungal resistance.
Keywords: Candida albicans; anaerobic; transcriptomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Zeise KD, Woods RJ, Huffnagle GB. 2021. Interplay between Candida albicans and lactic acid bacteria in the gastrointestinal tract: impact on colonization resistance, microbial carriage, opportunistic infection, and host immunity. Clin Microbiol Rev 34:e0032320. doi: 10.1128/CMR.00323-20 - DOI - PMC - PubMed
-
- Pradhan A, Avelar GM, Bain JM, Childers DS, Larcombe DE, Netea MG, Shekhova E, Munro CA, Brown GD, Erwig LP, Gow NAR, Brown AJP. 2018. Hypoxia promotes immune evasion by triggering β-glucan masking on the Candida albicans cell surface via mitochondrial and cAMP-protein kinase A signaling. MBio 9:e01318-18. doi: 10.1128/mBio.01318-18 - DOI - PMC - PubMed
-
- Savage HP, Bays DJ, Tiffany CR, Gonzalez MAF, Bejarano EJ, Carvalho TP, Luo Z, Masson HLP, Nguyen H, Santos RL, Reagan KL, Thompson GR, Bäumler AJ. 2024. Epithelial hypoxia maintains colonization resistance against Candida albicans. Cell Host Microbe 32:1103–1113. doi: 10.1016/j.chom.2024.05.008 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

