Genomic characterization of plasmids harboring blaNDM-1, blaNDM-5, and blaNDM-7 carbapenemase alleles in clinical Klebsiella pneumoniae in Pakistan
- PMID: 40401976
- PMCID: PMC12210987
- DOI: 10.1128/spectrum.02359-24
Genomic characterization of plasmids harboring blaNDM-1, blaNDM-5, and blaNDM-7 carbapenemase alleles in clinical Klebsiella pneumoniae in Pakistan
Abstract
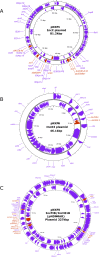
Klebsiella pneumoniae is notorious for causing healthcare-associated infections, which become more complicated by the acquisition of blaNDM genes via mobile genetic elements. Although Pakistan is a well-established hot spot of blaNDM-positive K. pneumoniae, detailed molecular descriptions of blaNDM-carrying plasmids are scarce. Seven K. pneumoniae isolates harboring blaNDM were recovered from clinical sample sources during a 6 month period and tested for antimicrobial susceptibility. A long-read approach was used for whole-genome sequencing to obtain circularized plasmids and chromosomes for typing, annotation, and comparative analysis. The isolates were susceptible to colistin and tigecycline only among the tested antibiotics. We identified five sequence types (STs): ST11, ST16, ST716, ST464, and ST2856. Notably, three strains possessed the hypervirulent capsule KL2, while five were classified as O locus type O2a. Evidence of genetic diversity was further highlighted by the presence of four IncC plasmids harboring blaNDM-1, two IncX3 plasmids harboring blaNDM-5, and a single hybrid IncFIB/IncHI1B plasmid harboring blaNDM-7. These plasmids also carried additional antimicrobial resistance (AMR) genes conferring resistance to aminoglycosides, cephalosporins, and fluoroquinolones. We identified the plasmidome of the K. pneumoniae isolates and characterized the New Delhi metallo-beta-lactamase (NDM)-carrying plasmids. Genetic analysis confirmed the presence of blaNDM-1 and blaNDM-5 on broad host range plasmids and blaNDM-7 in a previously unreported hybrid plasmid backbone. We emphasized the critical role of plasmids in spreading blaNDM in the clinical setting in Pakistan. Hence, we stressed the urgent need for enhanced surveillance, not least in low-middle income countries, infection control measures, and adherence to the "Access," "Watch," and "Reserve" guidelines in antibiotics use.
Importance: Infections caused by NDM-producing Klebsiella pneumoniae are a significant challenge to treat and represent a crucial health burden in low- and middle-income countries (LMICs). Most of the blaNDM are located on plasmids that promote horizontal gene transfer. However, there is a lack of comprehensive information on the genetic context of the NDM-carrying plasmids in Pakistan. This study presents a detailed analysis of seven NDM-plasmids in clinical K. pneumoniae isolates, shedding light on their high-risk sequence types and multiple resistance determinants. We also describe the plasmid-bearing NDM alleles (blaNDM-1, blaNDM-5, and blaNDM-7). Notably, we are the first to report blaNDM-7 on the hybrid IncFIB/IncHI1B backbone in Pakistan, a plasmid that has rarely been reported previously globally. Understanding the plasmid genomic landscape is paramount to comprehensively understanding the AMR scenario in this LMIC.
Keywords: CR-KP; NDM; antibiotic resistance; carbapenemases; mobile genetic elements; plasmids.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- van Duin D, Arias CA, Komarow L, Chen L, Hanson BM, Weston G, Cober E, Garner OB, Jacob JT, Satlin MJ, et al. 2020. Molecular and clinical epidemiology of carbapenem-resistant Enterobacterales in the USA (CRACKLE-2): a prospective cohort study. Lancet Infect Dis 20:731–741. doi: 10.1016/S1473-3099(19)30755-8 - DOI - PMC - PubMed
-
- Andrey DO, Pereira Dantas P, Martins WBS, Marques De Carvalho F, Almeida LGP, Sands K, Portal E, Sauser J, Cayô R, Nicolas MF, Vasconcelos ATR, Medeiros EA, Walsh TR, Gales AC. 2020. An emerging clone, Klebsiellapneumoniae carbapenemase 2–producing K. pneumoniae sequence type 16, associated with high mortality rates in a CC258-endemic setting. Clin Infect Dis 71:e141–e150. doi: 10.1093/cid/ciz1095 - DOI - PMC - PubMed
-
- WHO . 2024. WHO Bacterial Priority Pathogens List, 2024: bacterial pathogens of public health importance to guide research, development and strategies to prevent and control antimicrobial resistance - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

