A CARMIL2 gain-of-function mutation suffices to trigger most CD28 costimulatory functions in vivo
- PMID: 40402149
- PMCID: PMC12097149
- DOI: 10.1084/jem.20250339
A CARMIL2 gain-of-function mutation suffices to trigger most CD28 costimulatory functions in vivo
Abstract
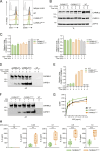
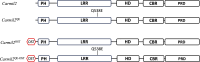
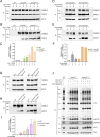
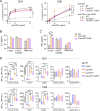
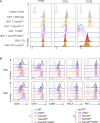
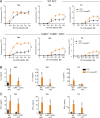
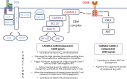
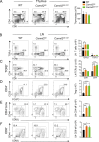
Naive T cell activation requires both TCR and CD28 signals. The CARMIL2 cytosolic protein enables CD28-dependent activation of the NF-κB transcription factor via its ability to link CD28 to the CARD11 adaptor protein. Here, we developed mice expressing a mutation named Carmil2QE and mimicking a mutation found in human T cell malignancies. Naive T cells from Carmil2QE mice contained preformed CARMIL2QE-CARD11 complexes in numbers comparable to those assembling in wild-type T cells after CD28 engagement. Such ready-made CARMIL2QE-CARD11 complexes also formed in CD28-deficient mice where they unexpectedly induced most of the functions that normally result from CD28 engagement in a manner that remains antigen-dependent. In turn, tumor-specific T cells expressing Carmil2QE do not require CD28 engagement and thereby escape to both PD-1 and CTLA-4 inhibition. In conclusion, we uncovered the overarching role played by CARMIL2-CARD11 signals among those triggered by CD28 and exploited them to induce potent solid tumor-specific T cell responses in the absence of CD28 ligands and immune checkpoint inhibitors.
© 2025 Zhang et al.
Conflict of interest statement
Disclosures: J. Celis-Gutierrez, R. Roncagalli, M. Malissen, and B. Malissen reported patent no. 23315449.1 (European Patent Office) pending. In addition, B. Malissen reported personal fees from AltruBio and personal fees from Janvier Group Biosciences outside the submitted work. No other disclosures were reported.
Figures






References
-
- Agarwal, S., Aznar M.A., Rech A.J., Good C.R., Kuramitsu S., Da T., Gohil M., Chen L., Hong S.A., Ravikumar P., et al. 2023. Deletion of the inhibitory co-receptor CTLA-4 enhances and invigorates chimeric antigen receptor T cells. Immunity. 56:2388–2407.e9. 10.1016/j.immuni.2023.09.001 - DOI - PMC - PubMed
-
- Ardouin, L., Luche H., Chelbi R., Carpentier S., Shawket A., Montanana Sanchis F., Santa Maria C., Grenot P., Alexandre Y., Grégoire C., et al. 2016. Broad and largely concordant molecular changes characterize tolerogenic and immunogenic dendritic cell maturation in thymus and periphery. Immunity. 45:305–318. 10.1016/j.immuni.2016.07.019 - DOI - PubMed
MeSH terms
Substances
Grants and funding
- 787300/ERC_/European Research Council/International
- Centre National de la Recherche Scientifique
- Institut National de la Santé et de la Recherche Médicale
- ANR-AAP-2018 Super-Basilic 18-CE15-0021-01/Agence Nationale de la Recherche
- ANR-AAP-2021 Carmil2 21-CE15-0034-02/Agence Nationale de la Recherche
- 32070898/National Natural Science Foundation of China
- 32170879/National Natural Science Foundation of China
- U1904157/National Natural Science Foundation of China
- 32000627/National Natural Science Foundation of China
- C24185/Henan Province
- D20036/National Discipline Innovation and Intelligence Introduction Base 111 Project
- ANR-10-INBS-08/Investissement d'Avenir program ProFi
- ANR-10-INBS-07/PHENOMIN
- DCBIOL
- Aix-Marseille Université
- MSDAvenir
LinkOut - more resources
Full Text Sources

