BRD9 functions as an HIV-1 latency regulatory factor
- PMID: 40402245
- PMCID: PMC12130862
- DOI: 10.1073/pnas.2418467122
BRD9 functions as an HIV-1 latency regulatory factor
Abstract
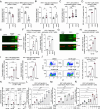
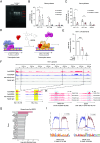
A major challenge for HIV type 1 (HIV-1) cure is the presence of viral latent reservoirs. The "Shock & Kill" strategy involves the combined use of latency reversal agents (LRA) and antiretroviral treatment (ART) to reactivate HIV-1 latent reservoirs, followed by elimination of infected cells. However, current LRAs are insufficient in fully reactivating the latent reservoirs. Therefore, investigation on novel HIV-1 latency regulators will be crucial to the success of HIV-1 cure research. Here, we identify bromodomain-containing protein 9 (BRD9) as an HIV-1 latency regulator. BRD9 inhibition induces HIV-1 latency reactivation in T cell lines, human resting memory CD4+ T cells, and PBMCs derived from people living with HIV-1 (PWH) on ART. BRD9 inhibition, gene depletion, and protein degradation consistently reactivate HIV-1 latency. Moreover, BRD9 inhibition synergizes with BRD4 inhibition in inducing HIV-1 production. Mechanistically, BRD9 binds to HIV-1 LTR promoter and competes with HIV-1 Tat protein for binding to the HIV-1 genome. Additionally, our integrated CUT&RUN DNA sequencing, transcriptomics, and pharmacological analysis revealed downstream host targets of BRD9, including ATAD2 and MTHFD2, that modulate HIV-1 latency.
Keywords: AIDS; BRD9; HIV-1 latency.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures



References
-
- Wong J. K., et al. , Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278, 1291–1295 (1997). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

