Emerging Technologies and Future Directions in Interorgan Crosstalk Cardiometabolic Research
- PMID: 40403107
- PMCID: PMC12101523
- DOI: 10.1161/CIRCRESAHA.125.325515
Emerging Technologies and Future Directions in Interorgan Crosstalk Cardiometabolic Research
Abstract
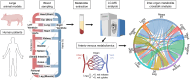
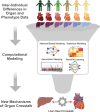
The heart does not work in isolation, with cardiac health and disease occurring through complex interactions between the heart with multiple organs. Furthermore, the integration of organ-specific lipid metabolism, blood pressure, insulin sensitivity, and inflammation involves a complex network of signaling pathways between many organs. Dysregulation in these communications is now recognized as a key contributor to many manifestations of cardiovascular disease. Mechanistic characterization of specific molecules mediating interorgan signaling has been pivotal in advancing our understanding of cardiovascular disease. The discovery of insulin, glucagon, and other hormones in the early 20th century illustrated the importance of communication between organs in maintaining physiological homeostasis. For example, elegant studies evaluating insulin signaling and its role in regulating glucose metabolism have shed light on its broader impact on cardiovascular health, hypertension, atherosclerosis, and other cardiovascular disease risks. Recent technological advances have revolutionized our understanding of interorgan signaling. Global approaches such as proteomics and metabolomics applications to blood have enabled the simultaneous profiling of thousands of circulating factors, revealing previously unknown signaling molecules and pathways. These large-scale studies have identified biomarkers linked to early stages of heart disease and offered new therapeutic targets. By understanding how specific cells in the heart interact with cells in other organs, such as the kidney or liver, researchers can identify key pathways that, when disrupted, lead to cardiovascular pathology. The ability to capture a more holistic view of the cardiovascular system positions interorgan signaling at the forefront of cardiovascular research. As we continue to refine our tools for mapping these complex networks, the insights gained hold the potential to not only improve early diagnosis but also to develop more targeted and effective treatments for cardiovascular disease. In this review, we discuss current approaches used to enhance our understanding of organ crosstalk with a specific emphasis on cardiac and cardiovascular physiology.
Keywords: blood pressure; cardiovascular diseases; cell communication; endocrine system; heart diseases; hypertension.
Conflict of interest statement
None.
Figures

Similar articles
-
Understanding different facets of cardiovascular diseases based on model systems to human studies: a proteomic and metabolomic perspective.J Proteomics. 2015 Sep 8;127(Pt A):50-60. doi: 10.1016/j.jprot.2015.04.027. Epub 2015 May 5. J Proteomics. 2015. PMID: 25956427 Review.
-
Proteomics Research in Cardiovascular Medicine and Biomarker Discovery.J Am Coll Cardiol. 2016 Dec 27;68(25):2819-2830. doi: 10.1016/j.jacc.2016.10.031. J Am Coll Cardiol. 2016. PMID: 28007144 Free PMC article. Review.
-
From Beats to Metabolism: the Heart at the Core of Interorgan Metabolic Cross Talk.Physiology (Bethesda). 2024 Mar 1;39(2):98-125. doi: 10.1152/physiol.00018.2023. Epub 2023 Dec 5. Physiology (Bethesda). 2024. PMID: 38051123 Review.
-
Proteomics and metabolomics combined in cardiovascular research.Trends Cardiovasc Med. 2007 Feb;17(2):43-8. doi: 10.1016/j.tcm.2006.11.004. Trends Cardiovasc Med. 2007. PMID: 17292045 Review.
-
Lipidomic insight into cardiovascular diseases.Biochem Biophys Res Commun. 2018 Oct 7;504(3):590-595. doi: 10.1016/j.bbrc.2018.04.106. Epub 2018 Apr 19. Biochem Biophys Res Commun. 2018. PMID: 29665359 Free PMC article. Review.
Cited by
-
Interorgan Crosstalk in Heart Failure and Cardiometabolic Diseases: A Compendium.Circ Res. 2025 May 23;136(11):1167-1169. doi: 10.1161/CIRCRESAHA.125.326720. Epub 2025 May 22. Circ Res. 2025. PMID: 40403104 Review. No abstract available.
References
-
- Lau WB, Ohashi K, Wang Y, Ogawa H, Murohara T, Ma X-L, Ouchi N. Role of adipokines in cardiovascular disease. Circ J. 2017;81:920–928. doi: 10.1253/circj.CJ-17-0458 - PubMed
-
- Shibata R, Ouchi N, Ohashi K, Murohara T. The role of adipokines in cardiovascular disease. J Cardiol. 2017;70:329–334. doi: 10.1016/j.jjcc.2017.02.006 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

