Expression characteristics of CsPG23 in citrus and analysis of its interacting protein
- PMID: 40403246
- PMCID: PMC12101599
- DOI: 10.1080/15592324.2025.2508418
Expression characteristics of CsPG23 in citrus and analysis of its interacting protein
Abstract
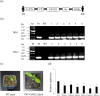
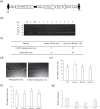
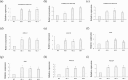
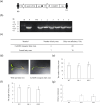
Exploring the resistance genes of citrus to Huanglongbing (HLB) is the foundation and key to citrus disease-resistant breeding. Through the analysis of comparative transcriptome data, we identified six cell wall degradation genes that respond to citrus infection with CaLas. We selected one of the genes with high differential expression levels and cloned it, naming it CsPG23. The subcellular localization results of tobacco indicated that the CsPG23 protein is localized in the nucleus, cytoplasm, and cell membrane. Real-time fluorescence quantitative PCR (RT-qPCR) analysis showed that the expression of CsPG23 is related to variety tolerance, tissue location, and symptom development. In addition, we constructed overexpression and silencing vectors for CsPG23 and obtained CsPG23 silencing plants, overexpression and silencing hairy roots, and analyzed the expression characteristics of CsPG23 in response to SA, JA, MeSA and H2O2 induction through RT-qPCR. Using Protein-Protein Interaction (PPI) to predict and screen for a citrus protein CsAGD8 that may interact with CsPG23, and preliminarily verifying its interaction with CsPG23 protein through Yeast Two-hybrid (Y2H). We constructed overexpression and silencing vectors for CsAGD8 and obtained CsAGD8 overexpression and silencing hairy roots. In summary, it is indicated that CsPG23 may interact with CsAGD8 in response to CaLas infection.
Keywords: Citrus; CsAGD8; CsPG23; hairy roots.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures






References
-
- Wulff NA, Zhang S, Setubal JC, Almeida NF, Martins EC, Harakava R, Kumar D, Rangel LT, Foissac X, Bové JM, et al. The complete genome sequence of ‘candidatus liberibacter americanus’, associated with citrus huanglongbing. Mol Plant-Microbe Interact: MPMI. 2014;27(2):163–176. doi: 10.1094/MPMI-09-13-0292-R. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
