BAF60/SWP73 subunits define subclasses of SWI/SNF chromatin remodelling complexes in Arabidopsis
- PMID: 40404167
- PMCID: PMC12177292
- DOI: 10.1111/nph.70182
BAF60/SWP73 subunits define subclasses of SWI/SNF chromatin remodelling complexes in Arabidopsis
Abstract
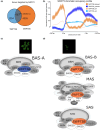
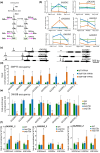
Evolutionarily conserved switch-defective/sucrose nonfermentable (SWI/SNF) ATP-dependent chromatin remodelling complexes (CRCs) alter nucleosome positioning and chromatin states, affecting gene expression to regulate important processes such as proper development and hormonal signalling pathways. We employed transcript profiling, chromatin immunoprecipitation (ChIP), mass spectrometry, yeast two-hybrid and bimolecular fluorescence complementation protein-protein interaction studies, along with hormone and metabolite profiling and phenotype assessments, to distinguish the SWP73A and SWP73B subunit functions in Arabidopsis. We identified a novel subclass of SWI/SNF CRCs defined by the presence of the SWP73A subunit. Therefore, we propose a refined classification of SWI/SNF CRCs in Arabidopsis, introducing BRM-associated SWI/SNF (BAS)-A (containing SWP73A) and BAS-B (containing SWP73B) subclasses. The SWP73A- and SWP73B-carrying SWI/SNF CRCs exhibit differential properties, demonstrated by distinct chromatin binding patterns and divergent effects on hormone biosynthesis and metabolism. We additionally found that SWP73A plays a specific role in the regulation of auxin signalling, root development, metabolism and germination that cannot be fully compensated by SWP73B. We recognised that some atypical subclasses of SWI/SNF CRCs may be likely formed in mutant lines with inactivated SWP73 subunits. Our study reveals that the duplication of the SWP73 subunit genes contributes to unique and shared functions of SWI/SNF CRC subclasses in the regulation of various processes in Arabidopsis.
Keywords: Arabidopsis; SWI/SNF; SWP73/BAF60; chromatin remodelling complexes; hormonal signalling; metabolome.
© 2025 The Author(s). New Phytologist © 2025 New Phytologist Foundation.
Conflict of interest statement
None declared.
Figures






References
-
- Alajem A, Biran A, Harikumar A, Sailaja BS, Aaronson Y, Livyatan I, Nissim‐Rafinia M, Sommer AG, Mostoslavsky G, Gerbasi VR et al. 2015. Differential association of chromatin proteins identifies BAF60a/SMARCD1 as a regulator of embryonic stem cell differentiation. Cell Reports 10: 2019–2031. - PubMed
-
- Archacki R, Buszewicz D, Sarnowski TJ, Sarnowska E, Rolicka AT, Tohge T, Fernie AR, Jikumaru Y, Kotlinski M, Iwanicka‐Nowicka R et al. 2013. BRAHMA ATPase of the SWI/SNF chromatin remodeling complex acts as a positive regulator of gibberellin‐mediated responses in Arabidopsis. PLoS ONE 8: e58588. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

