Single-cell RNA sequencing of baseline PBMCs predicts ICI efficacy and irAE severity in patients with NSCLC
- PMID: 40404203
- PMCID: PMC12097017
- DOI: 10.1136/jitc-2025-011636
Single-cell RNA sequencing of baseline PBMCs predicts ICI efficacy and irAE severity in patients with NSCLC
Abstract
Background: Immune checkpoint inhibitors (ICIs) have transformed treatment and have provided significant clinical benefits and durable responses for patients with advanced non-small cell lung cancer (NSCLC). However, only a small percentage of patients respond to ICI treatment, and immune-related adverse events (irAEs) leading to treatment discontinuation remain challenging. Despite the recognized need for biomarkers to predict both the efficacy of ICIs and the risk of irAEs, such biomarkers are yet to be clearly identified.
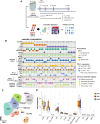
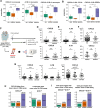
Methods: In this study, we performed single-cell RNA sequencing (scRNA-seq) of peripheral blood mononuclear cells (PBMCs) from 33 patients with NSCLC before ICIs treatment. To validate our findings, we reanalyzed public scRNA-seq data, conducted a cytometric bead array (CBA), and supported our findings with T-cell receptor sequencing.
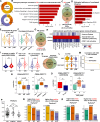
Results: While the immune response was more pronounced in patients with a favorable prognosis, the hypoxic pathway was more prominent in patients with primary resistance. Lymphocytes such as CD8 T cells, CD4 T cells, and natural killer cells were primarily involved in these pathways, with PRF1 and GZMB expression showing strong associations with favorable prognosis. In contrast, irAEs were mainly linked to myeloid cells, such as monocytes and macrophages. As irAE severity increased, inflammation and the TNF-NFKB1 pathway were more prominent. Specifically, increased expression of IL1B, CXCL8, and CXCL2 in monocytes and TNF in macrophages was closely associated with severe irAE through involvement in these pathways.Notably, the increase of PRF1 and GZMB expression showed a close association with both a favorable prognosis and a reduced severity of irAE, which was validated through CBA analysis. Moreover, the expression of these key markers varied according to prognosis and irAE severity regardless of patient background, such as programmed death-ligand 1 expression levels, tumor histology, or prior treatment regimens.
Conclusions: This study identified biological pathways and key biomarkers associated with ICI prognosis and irAE severity using PBMC samples before treatment. These findings provide a foundation for improved therapeutic strategies that enhance clinical outcomes while minimizing ICI treatment-associated risks.
Keywords: Biomarker; Immune Checkpoint Inhibitor; Immune related adverse event - irAE; Lung Cancer; Next generation sequencing - NGS.
© Author(s) (or their employer(s)) 2025. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ Group.
Conflict of interest statement
Competing interests: No, there are no competing interests.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
