Insights from the Biorepository and Integrative Genomics pediatric resource
- PMID: 40404628
- PMCID: PMC12098674
- DOI: 10.1038/s41467-025-59375-0
Insights from the Biorepository and Integrative Genomics pediatric resource
Abstract
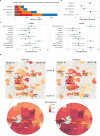
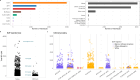
The Biorepository and Integrative Genomics (BIG) Initiative in Tennessee has developed a pioneering resource to address gaps in genomic research by linking genomic, phenotypic, and environmental data from a diverse Mid-South population, including underrepresented groups. We analyzed 13,152 exomes from BIG and found significant genetic diversity, with 50% of participants inferred to have non-European or several types of admixed ancestry. Ancestry within the BIG cohort is stratified, with distinct geographic and demographic patterns, as African ancestry is more common in urban areas, while European ancestry is more common in suburban regions. We observe ancestry-specific rates of novel genetic variants, which are enriched for functional or clinical relevance. Disease prevalence analysis linked ancestry and environmental factors, showing higher odds ratios for asthma and obesity in minority groups, particularly in the urban area. Finally, we observe discrepancies between self-reported race and genetic ancestry, with related individuals self-identifying in differing racial categories. These findings underscore the limitations of race as a biomedical variable. BIG has proven to be an effective model for community-centered precision medicine. We integrated genomics education, and fostered great trust among the contributing communities. Future goals include cohort expansion, and enhanced genomic analysis, to ensure equitable healthcare outcomes.
© 2025. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
Competing interests: The Regeneron Genetic Center is a subsidiary of Regeneron Pharmaceuticals, Inc. All the other authors declare no competing interests.
Figures


References
-
- Kyriazis, C. C. et al. Human genetic diversity and disease: from outside africa to within europe. Commun. Biol.6, 353 (2023).
-
- Sabeti, P. C. & Reich, D. Genetic and archeological evidence for early human population structure. Cell179, 1462–1474 (2019).
MeSH terms
LinkOut - more resources
Full Text Sources

