Kaposi's sarcoma-associated herpesvirus induces mitochondrial fission to evade host immune responses and promote viral production
- PMID: 40404827
- PMCID: PMC12337130
- DOI: 10.1038/s41564-025-02018-3
Kaposi's sarcoma-associated herpesvirus induces mitochondrial fission to evade host immune responses and promote viral production
Abstract
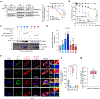
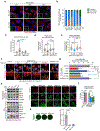
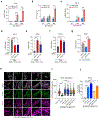
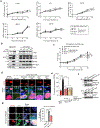
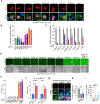
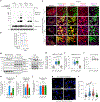
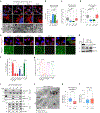
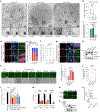
Mitochondrial dynamics are pivotal for host immune responses upon infection, yet how viruses manipulate these processes to impair host defence and enhance viral fitness remains unclear. Here we show that Kaposi's sarcoma-associated herpesvirus (KSHV), an oncogenic virus also known as human herpesvirus 8, encodes Bcl-2 (vBcl-2), which reprogrammes mitochondrial architecture. It binds with NM23-H2, a host nucleoside diphosphate (NDP) kinase, to stimulate GTP loading of the dynamin-related protein (DRP1) GTPase, which triggers mitochondrial fission, inhibits mitochondrial antiviral signalling protein (MAVS) aggregation and impairs interferon responses in cell lines. An NM23-H2-binding-defective vBcl-2 mutant fails to evoke fission, leading to defective virion assembly due to activated MAVS-IFN signalling. Notably, we identify two key interferon-stimulated genes restricting vBcl-2-dependent virion morphogenesis. Using a high-throughput drug screening, we discover an inhibitor targeting vBcl-2-NM23-H2 interaction that blocks virion production in vitro. Our study identifies a mechanism in which KSHV manipulates mitochondrial dynamics to allow for virus assembly and shows that targeting the virus-mitochondria interface represents a potential therapeutic strategy.
© 2025. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests: C.L., Q.Z. and R.M. are inventors of provisional patent 63/693,000, assigned to The Wistar Institute, and claiming compositions and methods disclosed herein. The other authors declare no competing interests.
Figures







References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

