Translocation of gut bacteria promotes tumor-associated mortality by inducing immune-activated renal damage
- PMID: 40404992
- PMCID: PMC12217037
- DOI: 10.1038/s44318-025-00458-5
Translocation of gut bacteria promotes tumor-associated mortality by inducing immune-activated renal damage
Abstract
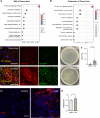
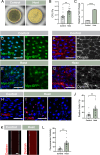
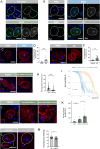
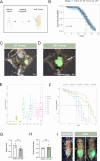
Paraneoplastic syndrome represents severe and complex systemic clinical symptoms manifesting in multiple organs of cancer patients, but its cause and cellular underpinnings remain little explored. In this study, establishing a Drosophila model of paraneoplastic syndrome triggered by tumor transplantation, we found that the innate immune response, initiated by translocated commensal bacteria from a compromised intestine, significantly contributes to reduced lifespan in tumor-bearing hosts. Our data identify the renal system as a central hub of this paraneoplastic syndrome model, wherein the pericardial nephrocytes undergo severe damage due to an elevated immune response triggered by gut dysbiosis and bacterial translocation. This innate immune response-induced nephrocyte damage is a major contributor to reduced longevity in tumor-bearing hosts, as blocking the NF-kB/Imd pathway in nephrocytes or removing gut bacteria via germ-free derivation or antibiotic treatment ameliorates nephrocyte deterioration and extends the lifespan of tumor-bearing flies. Consistently, treatment with a detoxifying drug also extended the lifespan of the tumor hosts. Our findings highlight a critical role of the gut-kidney axis in the paraneoplastic complications observed in cancer-bearing flies, suggesting potential therapeutic targets for mitigating similar complications in cancer patients.
Keywords: Drosophila Tumor Model; Bacterial Translocation; Gut-kidney Axis; Innate Immunity; Paraneoplastic Glomerulopathy.
© 2025. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures



References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

