Host genome drives the microbiota enrichment of beneficial microbes in shrimp: exploring the hologenome perspective
- PMID: 40405248
- PMCID: PMC12100935
- DOI: 10.1186/s42523-025-00414-y
Host genome drives the microbiota enrichment of beneficial microbes in shrimp: exploring the hologenome perspective
Abstract
Background: Pacific Whiteleg shrimp (Litopenaeus vannamei) is an important model for breeding programs to improve global aquaculture productivity. However, the interaction between host genetics and microbiota in enhancing productivity remains poorly understood. We investigated the effect of two shrimp genetic lines, Fast-Growth (Gen1) and Disease-Resistant (Gen2), on the microbiota of L. vannamei.
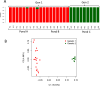
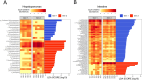
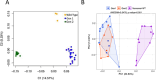
Results: Using genome-wide SNP microarray analysis, we confirmed that Gen1 and Gen2 represented distinct genetic populations. After confirming that the rearing pond did not significantly influence the microbiota composition, we determined that genetic differences explained 15.8% of the microbiota variability, with a stronger selective pressure in the hepatopancreas than in the intestine. Gen1, which exhibited better farm productivity, fostered a microbiota with greater richness, diversity, and resilience than Gen2, along with a higher abundance of beneficial microbes. Further, we demonstrated that a higher abundance of beneficial microbes was associated with healthier shrimp vs. diseased specimens, suggesting that Gen1 could improve shrimp's health and productivity by promoting beneficial microbes. Finally, we determined that the microbiota of both genetic lines was significantly different from their wild-type counterparts, suggesting farm environments and selective breeding programs strongly alter the natural microbiome.
Conclusions: This study highlights the importance of exploring the hologenome perspective, where integrating host genetics and microbiome composition can enhance breeding programs and farming practices.
Keywords: Breeding; Genetic line; Metagenomics; Microbiome; Microbiota-host interactions.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: An ethics statement was not required for the current study as locations for the specimen collection are not protected, and field studies did not include endangered or protected species. Animals were sacrificed under university protocols to avoid animal suffering. Competing interests: All authors declare no financial or non-financial competing interests.
Figures




Similar articles
-
Agavin induces beneficial microbes in the shrimp microbiota under farming conditions.Sci Rep. 2022 Apr 16;12(1):6392. doi: 10.1038/s41598-022-10442-2. Sci Rep. 2022. PMID: 35430601 Free PMC article.
-
Integrating short- and full-length 16S rRNA gene sequencing to elucidate microbiome profiles in Pacific white shrimp (Litopenaeus vannamei) ponds.Microbiol Spectr. 2024 Nov 5;12(11):e0096524. doi: 10.1128/spectrum.00965-24. Epub 2024 Sep 27. Microbiol Spectr. 2024. PMID: 39329828 Free PMC article.
-
Microbiome determinants of productivity in aquaculture of whiteleg shrimp.Appl Environ Microbiol. 2025 May 21;91(5):e0242024. doi: 10.1128/aem.02420-24. Epub 2025 Apr 15. Appl Environ Microbiol. 2025. PMID: 40231846 Free PMC article.
-
Understanding the role of the shrimp gut microbiome in health and disease.J Invertebr Pathol. 2021 Nov;186:107387. doi: 10.1016/j.jip.2020.107387. Epub 2020 Apr 21. J Invertebr Pathol. 2021. PMID: 32330478 Review.
-
The role of selective breeding and biosecurity in the prevention of disease in penaeid shrimp aquaculture.J Invertebr Pathol. 2012 Jun;110(2):247-50. doi: 10.1016/j.jip.2012.01.013. Epub 2012 Mar 13. J Invertebr Pathol. 2012. PMID: 22434005 Review.
References
-
- Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, et al. Host-gut microbiota metabolic interactions. Science. 2012;336(6086):1262–7. - PubMed
Grants and funding
- Fronteras de la Ciencia CF2019-G-263986/Consejo Nacional de Innovación, Ciencia y Tecnología
- Fronteras de la Ciencia CF2019-G-263986/Consejo Nacional de Innovación, Ciencia y Tecnología
- Fronteras de la Ciencia CF2019-G-263986/Consejo Nacional de Innovación, Ciencia y Tecnología
- Fronteras de la Ciencia CF2019-G-263986/Consejo Nacional de Innovación, Ciencia y Tecnología
- IN219723/Dirección General de Asuntos del Personal Académico (DGAPA)
LinkOut - more resources
Full Text Sources
