AP1-mediated reprogramming of EGFR expression triggers resistance to BLU-667 and LOXO-292 in RET-rearranged tumors
- PMID: 40405293
- PMCID: PMC12096543
- DOI: 10.1186/s13046-025-03392-w
AP1-mediated reprogramming of EGFR expression triggers resistance to BLU-667 and LOXO-292 in RET-rearranged tumors
Abstract
Background: Non-small cell lung cancer (NSCLC) is a significant global health challenge, with 2% of cases fuelled by RET rearrangements. RET inhibitors (RETi) have revolutionized treatment for these patients, but resistance remains an important clinical challenge limiting therapy effectiveness. This study investigated the mechanisms underlying resistance to RETi.
Methods: NSCLC cells were exposed to increasing doses of RETi (pralsetinib/BLU-667 and selpercatinib/LOXO-292) to generate resistant cells. RNA-Sequencing analysis identified differentially expressed genes in resistant versus sensitive cells, followed by in vitro and in vivo functional assays to explore novel therapeutic strategies. Additionally, tumor biopsies from RET-rearranged NSCLC patients who exhibited cancer progression on RET inhibitor therapy were analyzed.
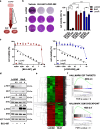
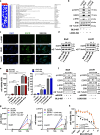
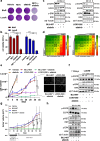
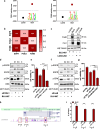
Results: RNA-sequencing analysis revealed the upregulation of the EGFR signaling pathway and hyperactivation of AP1 complex members in resistant cells compared to sensitive cells. Silencing of EGFR and AP1 complex members significantly reversed drug resistance, whereas EGFR overexpression reduced the sensitivity of parental Lc2/AD cells to RET inhibitors. Furthermore, the combination of RET and EGFR inhibitors showed synergistic antitumor activity in vitro and hindered tumor growth in mouse models with resistant cell xenografts. Notably, we observed a significant increase in EGFR expression in tumor biopsies from NSCLC patients treated with RET inhibitors who experienced disease progression, further validating the clinical relevance of our findings.
Conclusions: This study elucidates EGFR's role in mediating resistance to RET inhibitors in NSCLC patients. These findings offer insights into therapeutic adaptation and explore personalized combinations of RET and EGFR inhibitors for improved clinical outcomes.
Keywords: Combination targeted therapy; Drug resistance; EGFR; Non-small cell lung cancer; RET; RET inhibitors.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This study was reviewed and approved by the Federico II University Ethics Committee (ref n 139/22). Participants gave informed consent to participate in the study before taking part. The animal study was performed in accordance with institutional guidelines of the University of Naples Animal Care Committee and in accordance with the Declaration of Helsinki. Consent for publication: Not Applicable. Competing interests: Formisano declares the following competing interests: Consultant and advisory board for Seagen, Amgen, BMS, MSD, Jansen, and Pierre Fabre Pharma. Bianco declares the following competing interests: consultant and advisory board for BMS, MSD, Pfizer, AstraZeneca, Lilly and Novartis. Servetto reports honoraria from Eli Lilly, MSD, and Janssen and travel support from Bristol-Myers Squibb and AstraZeneca. Giuseppe Lo Russo declares advisory boards, consultancies, travel accommodations, speaker fees, writing fees, pi role in profit trials: MSD, Regeneron, Roche, Lilly, BMS, Amgen, AstraZeneca, Johnson And Johnson, Merck, Novartis, Pierre Fabre, Bayer, Beigene, Pfizer, Takeda, GSK, Daiichi,Sanofi, Gilead. Besse reports sponsored research at Gustave Roussy Cancer Center from AbbVie, Amgen, AstraZeneca, Biogen, Blueprint Medicines, Bristol-Myers Squibb, Celgene, Eli Lilly, GlaxoSmithKline, Ignyta, IPSEN, Merck KGaA, Merck Sharp & Dohme, Nektar, Onxeo, Pfizer, Pharma Mar, Sanofi, Spectrum Pharmaceuticals, Takeda, and Tiziana Pharma and investigators or coinvestigators of trials for Nerviano, GlaxoSmithKline, Pfizer, Roche-Genentech, Eli Lilly, OSE Pharma, Merck Sharp & Dohme, Celgene, Stemcentrx, Ignyta, AbbVie, Loxo Oncology, AstraZeneca, and Blueprint Medicines. Aldea reports receiving expenses and research funding from Sandoz and research funding from Amgen and AstraZeneca. Marinello declares travel/accommodation from Daiichi Sankyo and not financial support from MSD. The remaining authors declare that they have no competing interests.
Figures
References
-
- Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73:17–48. Available from: https://acsjournals.onlinelibrary.wiley.com/doi/10.3322/caac.21763. - DOI - PubMed
-
- Planchard D, Popat S, Kerr K, Novello S, Smit EF, Faivre-Finn C, et al. Metastatic non-small cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2018;29:iv192–237. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0923753419317107. - DOI - PubMed
-
- Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–50. Available from: https://www.nature.com/articles/nature13385. - PMC - PubMed
-
- Servetto A, Esposito D, Ferrara R, Signorelli D, Belli S, Napolitano F, et al. RET rearrangements in non-small cell lung cancer: Evolving treatment landscape and future challenges. Biochim Biophys Acta - Rev Cancer. 2022;1877:188810. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0304419X22001354. - DOI - PubMed
-
- Ju YS, Lee W-C, Shin JY, Lee S, Bleazard T, Won JK, et al. A transforming KIF5B and RET gene fusion in lung adenocarcinoma revealed from whole-genome and transcriptome sequencing. Genome Res. 2012;22:436–45. Available from: http://genome.cshlp.org/lookup/doi/10.1101/gr.133645.111. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- IG 21339/Associazione Italiana per la Ricerca sul Cancro
- MFAG 21505 - 2018/Associazione Italiana per la Ricerca sul Cancro
- MFAG 27826-2022/Associazione Italiana per la Ricerca sul Cancro
- IG20583/Associazione Italiana per la Ricerca sul Cancro
- Fellowship for Italy 2021 - 26795/Associazione Italiana per la Ricerca sul Cancro
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

