miRNA panel from HER2+ and CD24+ plasma extracellular vesicle subpopulations as biomarkers of early-stage breast cancer
- PMID: 40405296
- PMCID: PMC12096773
- DOI: 10.1186/s13058-025-02029-2
miRNA panel from HER2+ and CD24+ plasma extracellular vesicle subpopulations as biomarkers of early-stage breast cancer
Abstract
Background: Mammography screening has improved early breast cancer detection, leading to reduced mortality and lower rates of advanced breast cancer. However, mammography has a high false positive rate that results in over a million invasive breast biopsies of benign lesions in the US each year. Therefore, there is a need for noninvasive, blood-based diagnostics that can accurately assess risk of malignancy for women with indeterminate lesions identified by mammography, such as BI-RADS category 4 breast lesions. The aim of this study is to identify biomarkers from multiplexed extracellular vesicle liquid biopsy that can accurately classify mammographically detected BI-RADS 4 lesions.
Methods: We analyzed plasma from 113 prospectively enrolled subjects with BI-RADS 4 breast lesions, including 86 women with benign lesions and 27 women with malignant lesions (including 12 with stage I invasive carcinoma and 14 with ductal carcinoma in situ). None of the invasive carcinomas were metastatic. From each plasma sample, we used track etched magnetic nanopore technology to separately isolate HER2 and CD24 expressing extracellular vesicles (EVs) and measured their miRNA cargo using next-generation sequencing. We evaluated the performance of EV-miRNA biomarkers for classifying malignancy and applied LASSO classification to identify a panel of four complementary EV miRNA biomarkers that we validated by qPCR.
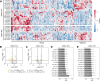
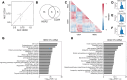
Results: We identified 19 differentially enriched miRNA from HER2+ EVs and 11 differentially enriched miRNA from CD24+ EVs of women with malignant lesions compared to benign lesions. We observed individual miRNA with an AUC of up to 0.87 for miR-340-5p from HER2+ EVs and 0.75 for miR-223-3p from CD24+ EVs. LASSO classification selected a panel of four complementary EV miRNA for classifying breast cancer: miR-340-5p (HER2+ EVs), miR-598-3p (CD24+), miR-15b-5p (HER2+), and miR-126-3p (CD24+).
Conclusions: HER2+ and CD24+ EV subpopulations contain complementary biomarkers suitable for validation in larger studies that can accurately detect early-stage breast cancer among women with BI-RADS category 4 breast lesions.
Keywords: BI-RADS 4 breast lesions; Early detection biomarkers; Extracellular vesicles; Liquid biopsy; MiRNA sequencing.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: All participants included in this study signed informed consent at the Hospital of the University of Pennsylvania (Philadelphia, PA) under IRB Protocol #833588. Consent for publication: Not applicable. Competing interests: DI holds ownership interest, including patents, in Chip Diagnostics. The other authors declare no competing interests.
Figures


References
-
- Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA: A Cancer Journal for Clinicians. 2023;73(1):17-48. - PubMed
-
- D'Orsi C, Sickles E, Mendelson E, Morris E. American College of Radiology Breast Imaging Reporting and Data System BI-RADS. Reston, VA2013.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

