TRPM8 levels determine tumor vulnerability to channel agonists
- PMID: 40405392
- PMCID: PMC12515718
- DOI: 10.1002/1878-0261.70049
TRPM8 levels determine tumor vulnerability to channel agonists
Abstract
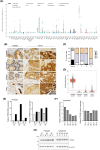
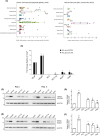
Targeted therapies have pervasively enhanced clinical protocols and significantly improved survival and quality of life of cancer patients. Mostly grounded on small molecules and antibodies targeting deregulated mechanisms in cancer cells, precision oncology approaches are limited to a few tumor types because of the paucity of clinically actionable targets. Here, we report a comparative analysis of the cation channel transient receptor potential melastatin 8 (TRPM8; also known as transient receptor potential cation channel subfamily M member 8) in lung, breast, colorectal, and prostate cancers. Our findings reveal high levels of channel expression in cores of all four carcinomas, irrespective of reduced expression of its RNA. Importantly, cancer cell lines that represent the various tumor types consistently show that sub-lethal chemotherapy dosages combined with the TRPM8 agonist D-3263 have a synergistic lethal effect. In addition, administration of D-3263 increases the cytotoxicity of 5-FU/Oxaliplatin in patient-derived colorectal cancer organoids, depending on the levels of TRPM8. Overall, our study strengthens the candidacy of TRPM8 as a molecular target for precision oncology approaches and paves the way for the design of basket trials for its clinical testing in TRPM8-high tumors.
Keywords: D‐3263; TRPM8; breast cancer; colorectal cancer; ion channel; lung cancer; prostate cancer.
© 2025 The Author(s). Molecular Oncology published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
S. Arena (SA) reports personal fees from MSD Italia and a patent (Italian patent application No. 102022000007535) outside the submitted work. A. Bardelli (ABa) declares the following competing financial interests: receipt of grants/research support from Neophore, AstraZeneca, Boehringer; receipt of honoraria or consultation fees from Guardant Health; stock shareholder: Neophore, Kither Biotech; member of the SAB of Neophore. No disclosures were reported by the other authors.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

