RaPID discovery of cell-permeable helical peptide inhibitors con-taining cyclic β-amino acids against SARS-CoV-2 main protease
- PMID: 40406165
- PMCID: PMC12093385
- DOI: 10.1039/d5cb00021a
RaPID discovery of cell-permeable helical peptide inhibitors con-taining cyclic β-amino acids against SARS-CoV-2 main protease
Abstract
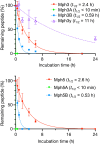
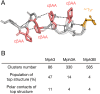
Structurally constrained cyclic β-amino acids are attractive building blocks for peptide drugs because they induce unique and stable conformations. Introduction of (1S,2S)-2-aminocyclopentanecarboxylic acid [(1S,2S)-2-ACPC] into peptides stabilizes helical conformations, so improving proteolytic stability and cell membrane permeability. We report on the ribosomal synthesis of a helical peptide library incorporating (1S,2S)-2-ACPC at every third position and its application for the discovery of SARS-CoV-2 main protease (Mpro) inhibitors. We identified two peptide sequences containing multiple (1S,2S)-2-ACPC residues, which exhibit helical conformations and superior proteolytic stability compared with their α-Ala or β-Ala counterparts. Studies using the chloroalkane cell-penetration assay showed that their cell permeability values (CP50) are comparable with or even slightly better than that of the cell-penetrating nona-arginine (R9) peptide. The new approach is thus a highly efficient method that combines a helical peptide library containing structurally constrained cyclic β-amino acids with the classical RaPID discovery method, enabling de novo discovery of proteolytically stable and cell-penetrating bioactive peptides that target intracellular proteins.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures




References
LinkOut - more resources
Full Text Sources
Miscellaneous

