Model systems to study Mycobacterium tuberculosis infections: an overview of scientific potential and impediments
- PMID: 40406522
- PMCID: PMC12095297
- DOI: 10.3389/fcimb.2025.1572547
Model systems to study Mycobacterium tuberculosis infections: an overview of scientific potential and impediments
Abstract
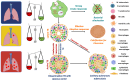
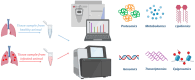
Despite years of global efforts to combat tuberculosis (TB), Mycobacterium tuberculosis (Mtb), the causative agent of this disease, continues to haunt the humankind making TB elimination a distant task. To comprehend the pathogenic nuances of this organism, various in vitro, ex vivo and in vivo experimental models have been employed by researchers. This review focuses on the salient features as well as pros and cons of various model systems employed for TB research. In vitro and ex vivo macrophage infection models have been extensively used for studying Mtb physiology. Animal models have provided us with great wealth of information and have immensely contributed to the understanding of TB pathogenesis and host responses during infection. Additionally, they have been used for evaluation of anti-mycobacterial drug therapy as well as for determining the efficacy of potential vaccine candidates. Advancements in various 'omics' based approaches have enhanced our understanding about the host-pathogen interface. Although animal models have been the cornerstone to TB research, none of them is ideal that gives us a complete picture of human infection, disease and progression. Further, the review also discusses about the newer systems including three dimensional (3D)-tissue models, lung-on-chip infection model, in vitro TB granuloma model and their limitations for studying TB. Thus, converging information gained from various in vitro and ex vivo models in tandem with in vivo experiments will ultimately bridge the gap that exists in understanding human TB.
Keywords: animal models; cellular models; host-pathogen interactions; omics-based approaches; tuberculosis.
Copyright © 2025 Nangpal, Nagpal, Angrish and Khare.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

