Alzheimer's Disease Sequencing Project release 4 whole genome sequencing dataset
- PMID: 40407102
- PMCID: PMC12100500
- DOI: 10.1002/alz.70237
Alzheimer's Disease Sequencing Project release 4 whole genome sequencing dataset
Abstract
Introduction: The Alzheimer's Disease Sequencing Project (ADSP) is a national initiative to understand the genetic architecture of Alzheimer's disease and related dementias (ADRD) by integrating whole genome sequencing (WGS) with other genetic, phenotypic, and harmonized datasets from diverse populations.
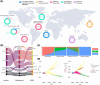
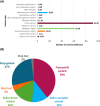
Methods: The Genome Center for Alzheimer's Disease (GCAD) uniformly processed WGS from 36,361 ADSP samples, including 35,014 genetically unique participants of which 45% are from non-European ancestry, across 17 cohorts in 14 countries in this fourth release (R4).
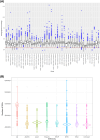
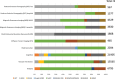
Results: This sequencing effort identified 387 million bi-allelic variants, 42 million short insertions/deletions, and 6.8 million structural variants. Annotations and quality control data are available for all variants and samples. Additionally, detailed phenotypes from 15,927 participants across 10 domains are also provided. A linkage disequilibrium panel was created using unrelated AD cases and controls.
Discussion: Researchers can access and analyze the genetic data via the National Institute on Aging Genetics of Alzheimer's Disease Data Storage Site (NIAGADS) Data Sharing Service, the VariXam, or NIAGADS GenomicsDB.
Highlights: We detailed the genetic architecture and quality of the Alzheimer's Disease Sequencing Project release 4 whole genome sequences. We identified 435 million single nucleotide polymorphisms, insertions and deletions, and structural variants from diverse genomes. We harmonized extensive phenotypes, linkage disequilibrium reference panel on subset of samples. Data is publicly available at NIAGADS Data Storage Site, variants and annotations are browsable on two different websites.
Keywords: Alzheimer's disease; diversity; genetic architecture; genetics data sharing; genetics knowledgebase; linkage disequilibrium reference panel; whole genome sequencing.
© 2025 The Author(s). Alzheimer's & Dementia published by Wiley Periodicals LLC on behalf of Alzheimer's Association.
Conflict of interest statement
G.S. receives payment or honoraria for lectures, presentations, speakers’ bureaus, manuscript writing, or educational events by BrightFocus and USC. T.J.H. serves on the scientific advisory board for Vivid Genomics, and as deputy editor for
Figures

Update of
-
Alzheimer's Disease Sequencing Project Release 4 Whole Genome Sequencing Dataset.medRxiv [Preprint]. 2024 Dec 6:2024.12.03.24317000. doi: 10.1101/2024.12.03.24317000. medRxiv. 2024. Update in: Alzheimers Dement. 2025 May;21(5):e70237. doi: 10.1002/alz.70237. PMID: 39677464 Free PMC article. Updated. Preprint.
Similar articles
-
Alzheimer's Disease Sequencing Project Release 4 Whole Genome Sequencing Dataset.medRxiv [Preprint]. 2024 Dec 6:2024.12.03.24317000. doi: 10.1101/2024.12.03.24317000. medRxiv. 2024. Update in: Alzheimers Dement. 2025 May;21(5):e70237. doi: 10.1002/alz.70237. PMID: 39677464 Free PMC article. Updated. Preprint.
-
NIAGADS: A data repository for Alzheimer's disease and related dementia genomics.Alzheimers Dement. 2025 Jun;21(6):e70255. doi: 10.1002/alz.70255. Alzheimers Dement. 2025. PMID: 40545618 Free PMC article.
-
Disentangling the genetic underpinnings of neuropsychiatric symptoms in Alzheimer's disease in the Alzheimer's Disease Sequencing Project: Study design and methodology.Alzheimers Dement (Amst). 2024 Aug 23;16(3):e70000. doi: 10.1002/dad2.70000. eCollection 2024 Jul-Sep. Alzheimers Dement (Amst). 2024. PMID: 39183746 Free PMC article.
-
18F PET with florbetapir for the early diagnosis of Alzheimer's disease dementia and other dementias in people with mild cognitive impairment (MCI).Cochrane Database Syst Rev. 2017 Nov 22;11(11):CD012216. doi: 10.1002/14651858.CD012216.pub2. Cochrane Database Syst Rev. 2017. PMID: 29164603 Free PMC article.
-
18F PET with flutemetamol for the early diagnosis of Alzheimer's disease dementia and other dementias in people with mild cognitive impairment (MCI).Cochrane Database Syst Rev. 2017 Nov 22;11(11):CD012884. doi: 10.1002/14651858.CD012884. Cochrane Database Syst Rev. 2017. PMID: 29164602 Free PMC article.
Cited by
-
Multiple-testing corrections in case-control studies using identity-by-descent segments.bioRxiv [Preprint]. 2025 Jul 7:2025.07.03.663057. doi: 10.1101/2025.07.03.663057. bioRxiv. 2025. PMID: 40672175 Free PMC article. Preprint.
-
Mosaic chromosomal alterations in blood are associated with an increased risk of Alzheimer's disease.medRxiv [Preprint]. 2025 Jun 4:2025.05.29.25328544. doi: 10.1101/2025.05.29.25328544. medRxiv. 2025. PMID: 40502569 Free PMC article. Preprint.
References
-
- Lambert J, Heath S, Even G, et al. Genome‐wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet. 2009;41(10):1094‐1099. - PubMed
MeSH terms
Grants and funding
- HHSN268201100012C/HL/NHLBI NIH HHS/United States
- Alzheimer's Therapeutic Research Institute
- P30 AG10161/Religious Orders Study
- NS039764/Mayo Parkinson's Disease
- P20 AG068082/AG/NIA NIH HHS/United States
- R01AG064614/Alzheimer's Therapeutic Researc Institute
- U01AG052410/Alzheimer's Therapeutic Researc Institute
- R01 AG042188/AG/NIA NIH HHS/United States
- P30 AG066444/AG/NIA NIH HHS/United States
- RC2 HL102419/HL/NHLBI NIH HHS/United States
- Austrian Stroke Prevention Study
- P30 AG013854/AG/NIA NIH HHS/United States
- P30 AG072975/AG/NIA NIH HHS/United States
- R37AG015473/Alzheimer's Disease Centers
- Alzheimer's Disease Sequencing Project (ADSP)
- Welfare and Sports
- U01AG057659/The Longitudinal Aging Study in India
- RC2AG036528/Arizona Department of Health Services
- R01AG057777/Darrell K Royal Texas Alzheimer's Initiative
- MRC_/Medical Research Council/United Kingdom
- R01 AG018016/AG/NIA NIH HHS/United States
- A2011048/University of Miami
- R01 AG060747/AG/NIA NIH HHS/United States
- 5R37AG015473/Estudio Familiar de Influencia Genetica en Alzheimer
- P01AG003991/Darrell K Royal Texas Alzheimer's Initiative
- HHSN268201100009I/HL/NHLBI NIH HHS/United States
- N01 HC085080/HL/NHLBI NIH HHS/United States
- R01 AG064877/AG/NIA NIH HHS/United States
- NS071674/Mayo Parkinson's Disease
- U01 AG046152/AG/NIA NIH HHS/United States
- P30 AG010124/AG/NIA NIH HHS/United States
- EuroImmun
- P30 AG072946/AG/NIA NIH HHS/United States
- R01AG051125/The Longitudinal Aging Study in India
- American Genome Center
- R01 NS017950/NS/NINDS NIH HHS/United States
- P30 AG066518/AG/NIA NIH HHS/United States
- R01 AG025711/Darrell K Royal Texas Alzheimer's Initiative
- Netherlands Consortium for Healthy Aging
- RC2 AG036528/AG/NIA NIH HHS/United States
- R01 AG019771/AG/NIA NIH HHS/United States
- P30 AG028377/AG/NIA NIH HHS/United States
- Alzheimer's Association "Identification of Rare Variants in Alzheimer Disease"
- Biogen
- P30 AG066507/AG/NIA NIH HHS/United States
- RF1AG054014/Arizona Department of Health Services
- U01 AG058654/AG/NIA NIH HHS/United States
- AG041718/Arizona Department of Health Services
- U01 HL096812/HL/NHLBI NIH HHS/United States
- R01 AG057777/AG/NIA NIH HHS/United States
- Johnson & Johnson Pharmaceutical Research & Development LLC.
- U54 AG052427/AG/NIA NIH HHS/United States
- Washington University Genome Institute
- R01AG058501/Darrell K Royal Texas Alzheimer's Initiative
- Framingham Heart Study
- R01 AG046949/AG/NIA NIH HHS/United States
- R01 AG042210/AG/NIA NIH HHS/United States
- P50 AG008671/AG/NIA NIH HHS/United States
- R01 AG054076/AG/NIA NIH HHS/United States
- U54 HG003067/HG/NHGRI NIH HHS/United States
- P50 AG005142/AG/NIA NIH HHS/United States
- R01 AG057909/AG/NIA NIH HHS/United States
- R01 AG058501/AG/NIA NIH HHS/United States
- 2014-A-004-NET/Hillblom Aging Network
- Ministry of Education
- U01AG062602/Gwangju Alzheimer and Related Dementias Study
- HHSN268201100010C/HL/NHLBI NIH HHS/United States
- RF1 AG057440/AG/NIA NIH HHS/United States
- European Special Populations Research Network
- R01AG044829/Longevity Genes Project
- U01 AG058922/AG/NIA NIH HHS/United States
- P20 AG068053/AG/NIA NIH HHS/United States
- R01 AG009029/AG/NIA NIH HHS/United States
- K08 AG034290/AG/NIA NIH HHS/United States
- R01 AG015928/AG/NIA NIH HHS/United States
- R01 AG044546/AG/NIA NIH HHS/United States
- P01 NS026630/NS/NINDS NIH HHS/United States
- P30 AG010133/AG/NIA NIH HHS/United States
- U24 AG021886/AG/NIA NIH HHS/United States
- Netherlands Organization for Scientific Research
- R01 AG009956/AG/NIA NIH HHS/United States
- HHSN268201100008C/HL/NHLBI NIH HHS/United States
- U01 HL080295/HL/NHLBI NIH HHS/United States
- P30 AG72975/Religious Orders Study
- IIRG09133827/University of Miami
- R01NS29993/Northern Manhattan Study
- Culture and Science
- 2R01AG09029/Multi-Institutional Research in Alzheimer's Genetic Epidemiology
- 5R37AG015473/Alzheimer's Therapeutic Researc Institute
- Victorian Forensic Institute of Medicine
- P50 AG005131/AG/NIA NIH HHS/United States
- P30AG038072/Longevity Genes Project
- R01 NS069719/NS/NINDS NIH HHS/United States
- U24AG056270/Alzheimer's Disease Centers
- CAPMC/ CIHR/Canada
- NU38CK000480/Case Western Reserve University Rapid Decline
- UF1AG047133/NH/NIH HHS/United States
- CurePSP Foundation
- HHSN268201500001C/HL/NHLBI NIH HHS/United States
- Reta Lila Weston Institute for Neurological Studies
- P50 AG005146/AG/NIA NIH HHS/United States
- Atherosclerosis Risk in Communities
- RF1AG053303/Alzheimer's Therapeutic Researc Institute
- K25 AG041906/AG/NIA NIH HHS/United States
- P30 AG066512/AG/NIA NIH HHS/United States
- U01 AG049507/AG/NIA NIH HHS/United States
- U01 HL096917/HL/NHLBI NIH HHS/United States
- P50 AG016574/AG/NIA NIH HHS/United States
- R01AG025259/Multi-Institutional Research in Alzheimer's Genetic Epidemiology
- P30 AG066511/AG/NIA NIH HHS/United States
- RF1AG053303/Darrell K Royal Texas Alzheimer's Initiative
- U24 AG072122/AG/NIA NIH HHS/United States
- Erasmus University
- HHSN268201100005G/HL/NHLBI NIH HHS/United States
- N01 HC085082/HL/NHLBI NIH HHS/United States
- R01 AG11101/Arizona Department of Health Services
- P01 AG017586/AG/NIA NIH HHS/United States
- RF1 AG054014/AG/NIA NIH HHS/United States
- HHSN268201100008I/HL/NHLBI NIH HHS/United States
- R01 AG15819/Religious Orders Study
- European Commission
- Alzheimer's Drug Discovery Foundation
- RF1AG058501/Darrell K Royal Texas Alzheimer's Initiative
- P50 AG05681/Darrell K Royal Texas Alzheimer's Initiative
- U01 AG052411/AG/NIA NIH HHS/United States
- R01AG064614/Darrell K Royal Texas Alzheimer's Initiative
- R01 AG019085/AG/NIA NIH HHS/United States
- Genetic and Environmental Risk Factors
- P30 AG066515/AG/NIA NIH HHS/United States
- RF1 AG053303/AG/NIA NIH HHS/United States
- U01 HL130114/HL/NHLBI NIH HHS/United States
- R01 AG017216/Darrell K Royal Texas Alzheimer's Initiative
- R01 AG11101/Chicago Health and Aging Project
- U01 AG032984/AG/NIA NIH HHS/United States
- R01AG032289/Hillblom Aging Network
- P30 AG062421/AG/NIA NIH HHS/United States
- R01 AG030146/AG/NIA NIH HHS/United States
- U01 AG046170/AG/NIA NIH HHS/United States
- R01 AG057907/AG/NIA NIH HHS/United States
- R01 AG09029/Multi-Institutional Research in Alzheimer's Genetic Epidemiology Study
- U01 AG024904/AG/NIA NIH HHS/United States
- ERC_/European Research Council/International
- N01 HC085086/HL/NHLBI NIH HHS/United States
- P50 AG008702/AG/NIA NIH HHS/United States
- N01 HC085083/HL/NHLBI NIH HHS/United States
- Lumosity
- R01AG028786/University of Miami Brain Endowment Bank
- HHSN268201100007C/HL/NHLBI NIH HHS/United States
- HHSN268200800007C/HL/NHLBI NIH HHS/United States
- R01 AG048234/AG/NIA NIH HHS/United States
- RF1AG015473/Estudio Familiar de Influencia Genetica en Alzheimer
- Ministry for Health
- U01AG065958/The Longitudinal Aging Study in India
- Piramal Imaging
- R01 AG043617/AG/NIA NIH HHS/United States
- Takeda Pharmaceutical Company
- U01 AG068057/AG/NIA NIH HHS/United States
- W81XWH-12-2-0012/National Institute on Aging Genetics of Alzheimer's Disease Data Storage Site
- Hussman Institute for Human Genomics
- U54HG003067/Broad Institute Genome Center
- P01AG017586/Tau Consortium
- HHSN268201100011I/HL/NHLBI NIH HHS/United States
- QLG2-CT-2002- 01254/Quality of Life and Management of the Living Resources
- HHSN268201100011C/HL/NHLBI NIH HHS/United States
- ALZ/Alzheimer's Association/United States
- RF1 AG054074/AG/NIA NIH HHS/United States
- Erasmus Rucphen Family Study
- AG064877/Arizona Department of Health Services
- U01 AG016976/AG/NIA NIH HHS/United States
- U54AG052427/Genome Center for Alzheimer's Disease
- P50 NS039764/NS/NINDS NIH HHS/United States
- P30AG066462/Darrell K Royal Texas Alzheimer's Initiative
- P30 AG066508/AG/NIA NIH HHS/United States
- R01 AG18023/Arizona Department of Health Services
- Genentech, Inc.
- R01 AG003949/Darrell K Royal Texas Alzheimer's Initiative
- P50 AG005681/AG/NIA NIH HHS/United States
- P01 AG003991/AG/NIA NIH HHS/United States
- Study Investigators institutions
- R56AG051876/Alzheimer's Therapeutic Researc Institute
- R01AG041797/University of Washington Families
- U24 AG056270/AG/NIA NIH HHS/United States
- P30 AG072978/AG/NIA NIH HHS/United States
- HEALTH-F4- 2007-201413/European Community's Seventh Framework
- P01 AG026276/AG/NIA NIH HHS/United States
- R01 AG017917/AG/NIA NIH HHS/United States
- RC2 HG005605/HG/NHGRI NIH HHS/United States
- U24 AG21886/The Longitudinal Aging Study in India
- R01AG046949/Longevity Genes Project
- RF1 AG058501/AG/NIA NIH HHS/United States
- P30 AG062429/AG/NIA NIH HHS/United States
- P01 AG03991/Darrell K Royal Texas Alzheimer's Initiative
- P30 AG19610/MND Victoria
- U01 HL096902/HL/NHLBI NIH HHS/United States
- P30 AG013846/AG/NIA NIH HHS/United States
- Rotterdam
- Netherlands Organization for Scientific Research and the Russian Foundation for Basic Research
- U01AG032984/Transition Therapeutics
- R56 AG051876/AG/NIA NIH HHS/United States
- U54NS100693/Arizona Department of Health Services
- R01AG064877/Darrell K Royal Texas Alzheimer's Initiative
- R01AG054047/Wisconsin Registry for Alzheimer's Prevention
- R01 AG028786/AG/NIA NIH HHS/United States
- Anniversary Fund
- U54 HG003273/HG/NHGRI NIH HHS/United States
- R01 AG049607/AG/NIA NIH HHS/United States
- Araclon Biotech
- P30 AG066519/AG/NIA NIH HHS/United States
- UF1 AG047133/AG/NIA NIH HHS/United States
- U01 AG057659/AG/NIA NIH HHS/United States
- KL2 RR024151/RR/NCRR NIH HHS/United States
- Medical University of Graz
- U24 AG074855/AG/NIA NIH HHS/United States
- R01 AG061155/AG/NIA NIH HHS/United States
- Novartis Pharmaceuticals Corporation
- P50 AG005136/AG/NIA NIH HHS/United States
- R01AG025259/Alzheimer Disease Among African Americans Study
- Meso Scale Diagnostics, LLC.
- CereSpir, Inc.
- P30 AG012300/AG/NIA NIH HHS/United States
- R01 AG42210/Religious Orders Study
- Northern California Institute for Research and Education
- Center for Genome Technology
- BioClinica, Inc.
- R01 AG027161/AG/NIA NIH HHS/United States
- RF1 AG054023/AG/NIA NIH HHS/United States
- R01 AG054047/AG/NIA NIH HHS/United States
- 5R01AG009956/Ibadan Study of Aging
- Meso Scale Diagnostics, LLC
- R01AG060747/Religious Orders Study
- U24AG041689/The Longitudinal Aging Study in India
- P30 AG072973/AG/NIA NIH HHS/United States
- U54AG052427/CurePSP
- P30 AG062422/AG/NIA NIH HHS/United States
- RF1 AG054080/AG/NIA NIH HHS/United States
- U19 AG024904/AG/NIA NIH HHS/United States
- U01 AG062943/AG/NIA NIH HHS/United States
- RF1 AG051504/AG/NIA NIH HHS/United States
- GE Healthcare
- P50 AG016573/AG/NIA NIH HHS/United States
- GHR Foundation
- R01 HL105756/HL/NHLBI NIH HHS/United States
- R01 AG079280/AG/NIA NIH HHS/United States
- Large Scale Sequencing and Analysis Centers
- K25 AG041906-01/Arizona Department of Health Services
- Research Institute for Diseases in the Elderly
- Austrian Science Fond
- RF1AG058501/Alzheimer's Disease Centers
- P30AG066444/Alzheimer's Therapeutic Researc Institute
- U.S. Department of Health and Human Services
- R01AG031272/Cache County Study
- Cardiovascular Health Study
- Medical Research Council UK
- RF1 AG058066/AG/NIA NIH HHS/United States
- Parkinson's Victoria
- R01AG042188/Longevity Genes Project
- Departments of Neurology and Psychiatry at Washington University School of Medicine
- 3U01AG052410/CubanAmerican Alzheimer's Disease Initiative
- U24 AG041689/AG/NIA NIH HHS/United States
- U01AG052410/University of Miami Brain Endowment Bank
- P50 AG005134/AG/NIA NIH HHS/United States
- U01 AG006781/NH/NIH HHS/United States
- RF1AG054080/Darrell K Royal Texas Alzheimer's Initiative
- P30 AG008017/AG/NIA NIH HHS/United States
- HHSN268201100006C/HL/NHLBI NIH HHS/United States
- R01AG044546/Alzheimer's Therapeutic Researc Institute
- R01 AG044829/AG/NIA NIH HHS/United States
- P30 AG066462/AG/NIA NIH HHS/United States
- R01NS069719/University of Washington Families
- U54AG052427/The Longitudinal Aging Study in India
- R01AG027161/Wisconsin Registry for Alzheimer's Prevention
- P30 AG010161/AG/NIA NIH HHS/United States
- P30 AG066530/AG/NIA NIH HHS/United States
- R01 AG033193/AG/NIA NIH HHS/United States
- R01AG057909/Longevity Genes Project
- Austrian National Bank
- HHSN268201200036C/HL/NHLBI NIH HHS/United States
- Victorian Brain Bank
- R01 AG036042/AG/NIA NIH HHS/United States
- P50 AG025688/AG/NIA NIH HHS/United States
- HHSN268201100005I/HL/NHLBI NIH HHS/United States
- R01 AG032990/AG/NIA NIH HHS/United States
- Netherlands Organization
- AbbVie
- 2R01AG048927/Multi-Institutional Research in Alzheimer's Genetic Epidemiology
- U01AG052410/Research in African American Alzheimer Disease Initiative
- P50AG008012/Case Western Reserve University Brain Bank
- Austrian Science Fund
- N01 HC055222/HL/NHLBI NIH HHS/United States
- U01 AG058589/AG/NIA NIH HHS/United States
- R37 AG015473/AG/NIA NIH HHS/United States
- R01AG21136/Cache County Study
- U01 NS041588/NS/NINDS NIH HHS/United States
- U01AG057659/Uniformed Services University of the Health Sciences
- R01 NS080820/NS/NINDS NIH HHS/United States
- Department of Internal Medicine
- R01 AG059716/AG/NIA NIH HHS/United States
- HHSN268201500001I/HL/NHLBI NIH HHS/United States
- P01AG03991/Alzheimer's Therapeutic Researc Institute
- U01 AG049508/AG/NIA NIH HHS/United States
- U01AG052410/Darrell K Royal Texas Alzheimer's Initiative
- P50 AG008012/AG/NIA NIH HHS/United States
- P01AG03991/Darrell K Royal Texas Alzheimer's Initiative
- P50 NS072187/NS/NINDS NIH HHS/United States
- U24AG041689/National Institute on Aging Genetics of Alzheimer's Disease Data Storage Site
- R01AG046170/Arizona Department of Health Services
- R01 AG15819/Arizona Department of Health Services
- Avid and Cogstate
- R01AG018016/Mexican Health and Aging Study
- RF1AG015473/Alzheimer's Therapeutic Researc Institute
- U01 HL096814/HL/NHLBI NIH HHS/United States
- Mayo Clinic Florida
- R01 AG074328/AG/NIA NIH HHS/United States
- U01AG032984/Arizona Department of Health Services
- RC2 AG036547/AG/NIA NIH HHS/United States
- Steiermärkische Krankenanstalten Gesellschaft
- U24AG041689/Transition Therapeutics
- N01 HC085079/HL/NHLBI NIH HHS/United States
- U01AG058922/Alzheimer's Therapeutic Researc Institute
- U01AG058922/Alzheimer's Disease Centers
- Cogstate
- U01 AG052410/AG/NIA NIH HHS/United States
- 5RC2HG005605/Mayo Parkinson's Disease
- P30 AG066509/AG/NIA NIH HHS/United States
- Erasmus MC
- U01 AG006781/AG/NIA NIH HHS/United States
- R01 AG041797/AG/NIA NIH HHS/United States
- EU Joint Programme - Neurodegenerative Disease Research
- EB/NIBIB NIH HHS/United States
- K23 AG030944/AG/NIA NIH HHS/United States
- U54NS100693/CurePSP
- P20 AG068077/AG/NIA NIH HHS/United States
- U01 AG062602/AG/NIA NIH HHS/United States
- R01 NS029993/NS/NINDS NIH HHS/United States
- R01AG044546/Darrell K Royal Texas Alzheimer's Initiative
- Johnson & Johnson Pharmaceutical Research & Development LLC
- RC4 AG039085/AG/NIA NIH HHS/United States
- Reasons for Geographic and Racial Differences in Stroke
- HHSN271201300031C/DA/NIDA NIH HHS/United States
- P30 AG066546/AG/NIA NIH HHS/United States
- P30AG066444/Darrell K Royal Texas Alzheimer's Initiative
- R01 AG064614/AG/NIA NIH HHS/United States
- BRIDGET
- P30 AG079280/Alzheimer's Disease Research Centers
- P30 AG038072/AG/NIA NIH HHS/United States
- Erasmus Medical Center
- R01 AG032289/AG/NIA NIH HHS/United States
- R01 AG048927/AG/NIA NIH HHS/United States
- U54 HG003079/HG/NHGRI NIH HHS/United States
- R01 AG019757/AG/NIA NIH HHS/United States
- U01 AG052409/AG/NIA NIH HHS/United States
- U01 AG046139/AG/NIA NIH HHS/United States
- R01 AG033040/AG/NIA NIH HHS/United States
- RF1 AG054052/AG/NIA NIH HHS/United States
- R01 AG007584/Genetic Differences
- R01 AG021547/AG/NIA NIH HHS/United States
- RF1AG054080/Alzheimer's Therapeutic Researc Institute
- R01AG11380/Cache County Study
- U01 AG006576/Darrell K Royal Texas Alzheimer's Initiative
- R01 AG051125/AG/NIA NIH HHS/United States
- Accelerating Medicines Partnership
- R56 AG064877/AG/NIA NIH HHS/United States
- P30 AG062677/AG/NIA NIH HHS/United States
- National Institutes of Health-National Institute on Aging
- Elan Pharmaceuticals, Inc.
- P30 AG072977/AG/NIA NIH HHS/United States
- R01AG064877/Alzheimer's Therapeutic Researc Institute
- R01 AG020098/AG/NIA NIH HHS/United States
- public-private-philanthropic partnership
- Rainwater Charitable Foundation
- RF1AG054074/Puerto Rican Alzheimer Disease Initiative
- Health Research and Development
- R01 HL070825/HL/NHLBI NIH HHS/United States
- RF1AG057440/Arizona Department of Health Services
- R01 AG027944/AG/NIA NIH HHS/United States
- HHSN268201100005C/HL/NHLBI NIH HHS/United States
- P30 AG072958/AG/NIA NIH HHS/United States
- R01 AG025259/AG/NIA NIH HHS/United States
- R01 AG030653/AG/NIA NIH HHS/United States
- HHSN268201100009C/HL/NHLBI NIH HHS/United States
- P20 AG068024/AG/NIA NIH HHS/United States
- Eli Lilly and Company
- P01 AG003949/AG/NIA NIH HHS/United States
- RF1 AG057519/AG/NIA NIH HHS/United States
- U01 HL096899/HL/NHLBI NIH HHS/United States
- P30 AG062715/AG/NIA NIH HHS/United States
- RF1 AG015473/AG/NIA NIH HHS/United States
- P50AG005136/University of Washington Families
- U01AG058922/Darrell K Royal Texas Alzheimer's Initiative
- Prospective Dementia Registry-Austria
- P30 AG072976/AG/NIA NIH HHS/United States
- P30 AG010129/AG/NIA NIH HHS/United States
- HHSN268201100007I/HL/NHLBI NIH HHS/United States
- R01AG064614/The Longitudinal Aging Study in India
- U01 AG049506/AG/NIA NIH HHS/United States
- U24 NS072026/NS/NINDS NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- R01 AG011101/AG/NIA NIH HHS/United States
- R01 AG17917/Arizona Department of Health Services
- U24 AG21886/Arizona Department of Health Services
- P30 AG066506/AG/NIA NIH HHS/United States
- U24-AG041689/University of Pennsylvania
- P30 AG066468/AG/NIA NIH HHS/United States
- R01 AG021136/AG/NIA NIH HHS/United States
- P30 AG019610/AG/NIA NIH HHS/United States
- IXICO Ltd.
- R01 AG041718/AG/NIA NIH HHS/United States
- Arizona Alzheimer's Disease Core Center
- UG3 NS104095/NS/NINDS NIH HHS/United States
- RF1AG058267/Case Western Reserve University Rapid Decline
- P30AG066462/Alzheimer's Therapeutic Researc Institute
- R01AG057907/Arizona Department of Health Services
- R01AG061155/Longevity Genes Project
- Hussman Institute for Human Genomics Brain Bank
- NeuroRx Research
- P30 AG072947/AG/NIA NIH HHS/United States
- P50 AG025711/AG/NIA NIH HHS/United States
- U01 AG058635/AG/NIA NIH HHS/United States
- Longitudinal Evaluation of Amyloid Risk and Neurodegeneration
- P30 AG072931/AG/NIA NIH HHS/United States
- Merck & Co., Inc.
- RF1AG053303/Alzheimer's Disease Centers
- RF1AG058066/Amish Protective Variant
- German Center for Neurodegenerative Diseases
- #NS072187/Morris K. Udall Parkinson's Disease Research Center of Excellence
- P30 AG072972/AG/NIA NIH HHS/United States
- U01 ES017155/ES/NIEHS NIH HHS/United States
- R01 AG30146/Arizona Department of Health Services
- R01 AG023629/AG/NIA NIH HHS/United States
- RF1AG054052/Cache County Study
- Janssen Alzheimer Immunotherapy Research & Development, LLC
- RF1AG058501/Alzheimer's Therapeutic Researc Institute
- R01AG064877/Alzheimer's Disease Centers
- P30 AG066514/AG/NIA NIH HHS/United States
- R56AG051876/Estudio Familiar de Influencia Genetica en Alzheimer
- U19 AG066567/NH/NIH HHS/United States
- U54AG052427/Transition Therapeutics
- P30 AG028383/AG/NIA NIH HHS/United States
- U19 AG066567/AG/NIA NIH HHS/United States
- P01 AG017216/AG/NIA NIH HHS/United States
- P30 AG072959/AG/NIA NIH HHS/United States
- P30-AG066468/Arizona Department of Health Services
- R01 AG031272/AG/NIA NIH HHS/United States
- Austrian Research Promotion agency
- P01AG026276/Darrell K Royal Texas Alzheimer's Initiative
- R01 AG011380/AG/NIA NIH HHS/United States
- Netherlands Genomics Initiative
- R01AG048234/Hillblom Aging Network
- P01AG026276/Alzheimer's Therapeutic Researc Institute
- P50 NS071674/NS/NINDS NIH HHS/United States
- R01 AG018023/AG/NIA NIH HHS/United States
- Neurotrack Technologies
- R01 AG17917/Memory and Aging Project
- Fujirebio
- AG030653/Arizona Department of Health Services
- Lundbeck
- N01 HC085081/HL/NHLBI NIH HHS/United States
- R01AG044546/Alzheimer's Disease Centers
- University of Toronto (UT)
- U01 AG006786/AG/NIA NIH HHS/United States
- U54 NS100693/NS/NINDS NIH HHS/United States
- 2R01AG09029/Alzheimer Disease Among African Americans Study
- Netherlands Organization of Scientific Research NWO Investments
- RF1AG054074/Peru Alzheimer's Disease Initiative
- Rotterdam Study
- P30 AG072979/AG/NIA NIH HHS/United States
- R01 AG015819/AG/NIA NIH HHS/United States
- R01 AG036836/AG/NIA NIH HHS/United States
- Eisai Inc.
- RF1 AG058267/AG/NIA NIH HHS/United States
- 2R01AG048927/Alzheimer Disease Among African Americans Study
- P30 AG10161/Arizona Department of Health Services
- U19AG024904)/Alzheimer's Disease Neuroimaging Initiative
- U54AG052427/Arizona Department of Health Services
- National Institute on Aging Alzheimer's Disease Data Storage Site
- U01 AG049505/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

