Acquisition of daptomycin resistance in patients results in decreased virulence in Drosophila
- PMID: 40407308
- PMCID: PMC12150764
- DOI: 10.1128/iai.00594-24
Acquisition of daptomycin resistance in patients results in decreased virulence in Drosophila
Abstract
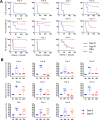
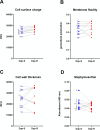
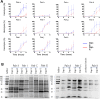
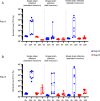
Staphylococcus aureus can acquire antimicrobial resistance, which in turn may affect its pathogenic potential. Using a panel of paired clinical isolates collected before and after daptomycin resistance acquisition, most frequently through a single mprF mutation, we show a relationship between increasing daptomycin minimum inhibitory concentration and reduced virulence in a Drosophila systemic infection model. Analyzing toxin production, in vitro bacterial growth characteristics, and cell surface properties, we failed to link daptomycin resistance-related attenuated virulence to either reduced virulence factor production, reduced fitness, or any of the cell surface characteristics investigated. Competition assays in Drosophila also did not support any altered ability in immune evasion. Instead, using a panel of mutant flies defective for various immune components, we show that this daptomycin resistance-related attenuated virulence is mostly explained by greater susceptibility to the activity of Drosophila prophenoloxidase, a tyrosinase involved in melanization, but not to antimicrobial peptides or Bomanin antimicrobial effectors. Further investigation could not link daptomycin resistance-related attenuation of virulence to differential susceptibility to reactive oxygen species or quinones prominently associated with phenoloxidase bacterial-killing activity. Taken together, it appears that daptomycin resistance attenuates Staphylococcus aureus virulence through enhanced sensitivity to phenoloxidase based on a complex mechanism. Our study provides new insights into the understanding of the crosstalk between antimicrobial resistance, escape from immune killing, and virulence.IMPORTANCEThis study advances current knowledge in the field of host-microbe interactions and antimicrobial resistance by exploring crosstalk between antimicrobial resistance and virulence. It shows how acquiring antimicrobial resistance can alter bacterial virulence and helps shape virulence. Relative to the parental staphylococcal strain, daptomycin-resistant clinical isolates most often varied by one single mutation in a gene involved in the composition of the bacterial membrane, and these strains were much less virulent when fruit flies were infected. The difference in virulence is unrelated to changes in bacterial toxin production, bacterial growth, immune evasion, or cell surface properties. Instead, resistant strains were more vulnerable to a host proenzyme involved in the antibacterial melanization response, an important response deployed throughout the arthropods. We predict that daptomycin resistance forces staphylococci to alter the composition of their cell surface, which causes the bacteria to become more vulnerable to killing by melanization.
Keywords: Drosophila melanogaster; Staphylococcus aureus; antimicrobial peptides; antimicrobial resistance; daptomycin; host-pathogen interaction; link between antimicrobial resistance and virulence; prophenoloxidase; virulence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Bai AD, Lo CKL, Komorowski AS, Suresh M, Guo K, Garg A, Tandon P, Senecal J, Del Corpo O, Stefanova I, Fogarty C, Butler-Laporte G, McDonald EG, Cheng MP, Morris AM, Loeb M, Lee TC. 2022. Staphylococcus aureus bacteraemia mortality: a systematic review and meta-analysis. Clin Microbiol Infect 28:1076–1084. doi: 10.1016/j.cmi.2022.03.015 - DOI - PubMed
-
- Le Moing V, Alla F, Doco-Lecompte T, Delahaye F, Piroth L, Chirouze C, Tattevin P, Lavigne J-P, Erpelding M-L, Hoen B, Vandenesch F, Duval X, VIRSTA study group . 2015. Staphylococcus aureus bloodstream infection and endocarditis - a prospective cohort study. PLoS One 10:e0127385. doi: 10.1371/journal.pone.0127385 - DOI - PMC - PubMed
-
- Habib G, Lancellotti P, Antunes MJ, Bongiorni MG, Casalta J-P, Del Zotti F, Dulgheru R, El Khoury G, Erba PA, Iung B, Miro JM, Mulder BJ, Plonska-Gosciniak E, Price S, Roos-Hesselink J, Snygg-Martin U, Thuny F, Tornos Mas P, Vilacosta I, Zamorano JL, ESC Scientific Document Group . 2015. 2015 ESC guidelines for the management of infective endocarditis: the task force for the management of infective endocarditis of the European Society of Cardiology (ESC)Endorsed by: European Association for Cardio-Thoracic Surgery (EACTS), the European Association of Nuclear Medicine (EANM). Eur Heart J 36:3075–3128. doi: 10.1093/eurheartj/ehv319 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

