Unraveling the molecular mechanisms of DNA capture by the Com pilus in naturally transformable monoderm bacteria
- PMID: 40407325
- PMCID: PMC12153270
- DOI: 10.1128/mbio.00851-25
Unraveling the molecular mechanisms of DNA capture by the Com pilus in naturally transformable monoderm bacteria
Abstract
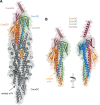
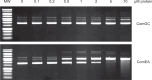
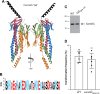
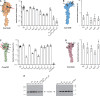
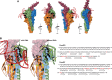
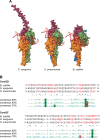
Transformation is a mechanism of horizontal gene transfer widespread in bacteria. The first step in transformation-capture of exogenous DNA-is mediated by surface-exposed filaments belonging to the type 4 filament (T4F) superfamily. How these protein polymers, composed of major and minor pilin subunits, interact with DNA remains poorly understood. Here, we address this question for the Com pilus, a widespread T4F mediating DNA capture in competent monoderm species. Our functional analysis, performed in Streptococcus sanguinis, was guided by a complete structural model of the Com pilus. We show that the major pilin ComGC does not bind DNA. In contrast, a systematic mutational analysis of electropositive residues exposed at the filament surface in the four minor pilins (ComGD, ComGE, ComGF, and ComGG) reveals that the interface between ComGD and ComGF is important for DNA capture. Sequential mutations in these two interacting subunits lead to complete abolition of transformation, without affecting piliation. We further demonstrate the physical interaction between ComGD and ComGF using disulfide crosslinking, upon mutagenesis of two strategically positioned residues into cysteines. A structural model of the Com pilus tip interacting with DNA recapitulates all these findings and highlights a novel mode of DNA-binding, conserved in hundreds of monoderm species.
Importance: Bacteria are capable of evolving and diversifying very rapidly by acquiring new genetic material via horizontal gene transfer (HGT). Transformation is a widespread mechanism of HGT, which results from the capture of extracellular DNA by surface-exposed pili belonging to the type 4 filament (T4F) superfamily. How T4F-composed of major and minor pilins-interact with DNA remains poorly understood, especially in monoderm species that use a unique T4F for DNA capture, known as Com pilus or T4dP. The significance of this work is in characterizing a novel mode of DNA-binding by showing that the interface between two minor pilins, part of a tip-located complex of four pilins-found in different T4F-has been functionalized in monoderms to capture DNA. This is an evolutionary mechanism promoting the exceptional functional versatility of T4F.
Keywords: DNA-binding proteins; genetic competence; gram-positive bacteria; natural transformation systems; type 4 pili.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

