Human endothelial colony forming cells (ECFCs) require endothelial protein C receptor (EPCR) for cell cycle progression and angiogenic activity
- PMID: 40407837
- PMCID: PMC12102113
- DOI: 10.1007/s10456-025-09982-8
Human endothelial colony forming cells (ECFCs) require endothelial protein C receptor (EPCR) for cell cycle progression and angiogenic activity
Abstract
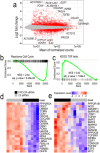
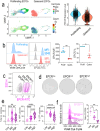
Vascular repair and regeneration are critical for tissue homeostasis. Endothelial colony forming cells (ECFCs) are vessel-resident progenitors with vasoreparative capacity and they offer an important avenue for allogeneic cytotherapy to achieve perfusion of ischemic tissues. Endothelial Protein C Receptor (EPCR) has been proposed as a marker for vascular endothelial stem cells, but its precise role in ECFC biology remains unknown. The current study has investigated the biological relevance of EPCR in ECFC function. Our data show that over 95% of ECFCs exhibit high EPCR expression. These levels surpassing CD34 and CD157, positions EPCR as a new robust ECFC immunophenotypic marker, alongside established markers CD31 and CD105. Functionally, depleting EPCR expression in ECFCs significantly diminished angiogenic activity, including proliferation, migration and tube formation. This knockdown also altered normal ECFC barrier function. Transcriptomic analysis indicated that knockdown of EPCR led to enrichment of gene signatures for cell cycle, TGF beta, and focal adhesion kinases. G1 cell cycle arrest was confirmed in ECFCs with depleted EPCR. Mechanistically, EPCR knockdown led to increased release of TGFβ2 and SMAD2/3 activation, coupled with increased p21, decreased pFAK, and increased transgelin. Additionally, we showed that quiescent ECFCs showed significantly lower EPCR expression when compared to proliferating ECFCs. In agreement with this, cell sorting experiments demonstrated that ECFCs with the highest EPCR expression exhibited the highest clonogenic capacity. In summary, our findings highlight that EPCR expression in ECFCs is critical for their angiogenic activity, by modulating cell cycle progression.
Keywords: Angiogenesis; Cell cycle; Endothelial colony forming cells; Endothelial progenitor; Protein C receptor; Transforming growth factor beta.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Conflict of interest: RJM and AWS are co-founders and scientific advisors for VascVersa Ltd. Ethical approval: The study has ethical approval by the NRES Committee with REC reference 15/YH/0281.
Figures




References
-
- Blandinières A, Randi AM, Paschalaki KE et al (2023) Results of an international survey about methods used to isolate human endothelial colony-forming cells: guidance from the SSC on Vascular Biology of the ISTH. J Thromb Haemost. 10.1016/j.jtha.2023.06.014 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

