Single-cell RNA sequencing and traditional RNA sequencing reveals the role of cancer-associated fibroblasts in head and neck squamous cell carcinomas cohort
- PMID: 40407978
- PMCID: PMC12102450
- DOI: 10.1007/s12672-025-02507-1
Single-cell RNA sequencing and traditional RNA sequencing reveals the role of cancer-associated fibroblasts in head and neck squamous cell carcinomas cohort
Abstract
Background: Head and neck squamous cell carcinomas (HNSCs) are among the most common tumors worldwide. Despite the availability of various diagnostic and therapeutic strategies, the incidence and mortality rates of HNSC remain high. Cancer-associated fibroblasts (CAFs), as a major component of the tumor microenvironment, exhibit diverse biological characteristics in terms of origin, genetics, and phenotype, and have been increasingly recognized for their roles in tumor progression.
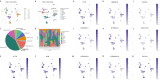
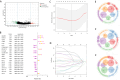
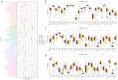
Methods: To investigate the potential role of CAFs in HNSC, we performed a comprehensive bioinformatics analysis based on the TCGA HNSC cohort. We applied single-sample gene set enrichment analysis (ssGSEA), single-cell RNA sequencing (scRNA-seq) analysis, differential expression analysis, Cox regression, LASSO regression, and pathway enrichment analysis to identify CAF-related genes and assess their prognostic value.
Results: We successfully identified a set of CAF-related genes and stratified the HNSC patients into high- and low-CAF groups. Based on the expression of these genes, we constructed a prognostic prediction model using LASSO and multivariate Cox regression analyses. A nomogram integrating the risk score and clinical characteristics was developed to improve individualized survival prediction. Enrichment analysis revealed that the type I interferon signaling pathway, cellular response to type I interferon, defense response to symbiont, and extracellular matrix organization were significantly associated with CAFs in HNSC.
Conclusion: Our study provides a novel CAF-based prognostic model and nomogram for predicting patient outcomes in HNSC. These findings highlight the importance of CAFs in the tumor microenvironment and their potential as therapeutic and prognostic biomarkers.
Keywords: Cancer-associated fibroblast; Head and neck squamous cell carcinoma; Immune cell infiltration; Single-cell RNA sequencing.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures





References
-
- Gu Z, Yao Y, Yang G, Zhu G, Tian Z, Wang R, Wu Q, Wang Y, Wu Y, Chen L, Wang C, Gao J, Kang X, Zhang J, Wang L, Duan S, Zhao Z, Zhang Z, Sun S. Pharmacogenomic landscape of head and neck squamous cell carcinoma informs precision oncology therapy. Sci Transl Med. 2022. 10.1126/scitranslmed.abo5987. - PubMed
-
- Moon DH, Sher DJ. Oligometastasis in head and neck squamous cell carcinoma. Int J Radiat Oncol Biol Phys. 2022;114(4):803–11. 10.1016/j.ijrobp.2022.06.086. - PubMed
-
- Harrington KJ, Burtness B, Greil R, Soulières D, Tahara M, de Castro G, Psyrri A, Brana I, Basté N, Neupane P, Bratland Å, Fuereder T, Hughes BGM, Mesia R, Ngamphaiboon N, Rordorf T, Wan Ishak WZ, Lin J, Gumuscu B, Swaby RF, Rischin D. Pembrolizumab with or without chemotherapy in recurrent or metastatic head and neck squamous cell carcinoma: updated results of the phase III KEYNOTE-048 study. J Clin Oncol. 2023;41(4):790–802. 10.1200/JCO.21.02508. - PMC - PubMed
LinkOut - more resources
Full Text Sources
