The type IV secretion system of Patescibacteria is homologous to the bacterial monoderm conjugation machinery
- PMID: 40408144
- PMCID: PMC12102498
- DOI: 10.1099/mgen.0.001409
The type IV secretion system of Patescibacteria is homologous to the bacterial monoderm conjugation machinery
Abstract
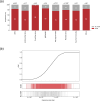
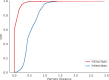
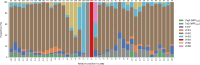
The Candidate Phyla Radiation, also known as Patescibacteria, represents a vast and diverse division of bacteria that has come to light via culture-independent 'omics' technologies. Their limited biosynthetic capacity, along with evidence of their growth as obligate epibionts on other bacteria, suggests a broad reliance on host organisms for their survival. Nevertheless, our understanding of the molecular mechanisms governing their metabolism and lifestyle remains limited. The type IV secretion system (T4SS) represents a superfamily of translocation systems with a wide range of functional roles. T4SS genes have been identified in the Patescibacteria class Saccharimonadia as essential for their epibiotic growth. In this study, we used a comprehensive bioinformatics approach to investigate the diversity and distribution of T4SS within Patescibacteria. The phylogenetic analysis of the T4SS signature protein VirB4 suggests that most of these proteins cluster into a distinct monophyletic group with a shared ancestry to the MPFFATA class of T4SS. This class is found in the conjugative elements of Firmicutes, Actinobacteria, Tenericutes and Archaea, indicating a possible horizontal gene transfer from these monoderm micro-organisms to Patescibacteria. We identified additional T4SS components near virB4, particularly those associated with the MPFFATA class, as well as homologues of other T4SS classes, such as VirB2-like pilins, and observed their varied arrangements across different Patescibacteria classes. The absence of a relaxase in most of these T4SS clusters suggests that the system has been co-opted for other functions in Patescibacteria. The proximity of T4SS components to the origin of replication (gene dnaA) in some Patescibacteria suggests a potential mechanism for increased expression. The broad ubiquity of a phylogenetically distinct T4SS, combined with its chromosomal location, underscores the significance of T4SS in the biology of Patescibacteria.
Keywords: Candidate Phyla Radiation; Patescibacteria; horizontal gene transfer; type IV secretion system.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

