Many transcription factor families have evolutionarily conserved binding motifs in plants
- PMID: 40408265
- PMCID: PMC12142463
- DOI: 10.1093/plphys/kiaf205
Many transcription factor families have evolutionarily conserved binding motifs in plants
Abstract
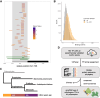
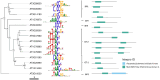
Transcription factors control gene expression during development and in response to a broad range of internal and external stimuli. They regulate promoter activity by directly binding cis-regulatory elements in DNA. The angiosperm Arabidopsis (Arabidopsis thaliana) contains more than 1,500 annotated transcription factors, each containing a DNA-binding domain that is used to define transcription factor families. Analyzing the binding motifs of 686 and the binding sites of 335 Arabidopsis transcription factors, as well as motifs of 92 transcription factors from other plants, we identified a constrained vocabulary of 74 conserved motifs spanning 50 families in plants. Among 21 transcription factor families, we found 1 core motif for all analyzed members and between 2% and 72% overlapping binding sites. Five families show conservation of the motif along phylogenetic clades. Five families, including the C2H2 zinc finger family, show high diversity among motifs in plants, suggesting potential for the neofunctionalization of duplicated transcription factors based on the motif recognized. We tested whether conserved motifs remained conserved since at least 450 million years ago by determining the binding motifs of 17 transcription factors from 11 families in Marchantia (Marchantia polymorpha) using amplified DNA affinity purification sequencing. We detected nearly identical binding motifs as predicted from the angiosperm data. Our findings show a large repertoire of overlapping binding sites within a transcription factor family and species and a high degree of binding motif conservation for at least 450 million years, indicating more potential for evolution in cis- rather than trans-regulatory elements.
© The Author(s) 2025. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Conflict of interest statement
Conflict of interest statement. None declared.
Figures



Similar articles
-
A conserved ARF-DNA interface underlies auxin-triggered transcriptional response.Proc Natl Acad Sci U S A. 2025 Apr 8;122(14):e2501915122. doi: 10.1073/pnas.2501915122. Epub 2025 Apr 1. Proc Natl Acad Sci U S A. 2025. PMID: 40168121 Free PMC article.
-
MpANT regulates meristem development in Marchantia polymorpha.Cell Rep. 2024 Jul 23;43(7):114466. doi: 10.1016/j.celrep.2024.114466. Epub 2024 Jul 9. Cell Rep. 2024. PMID: 38985681
-
Jasmonate-Related MYC Transcription Factors Are Functionally Conserved in Marchantia polymorpha.Plant Cell. 2019 Oct;31(10):2491-2509. doi: 10.1105/tpc.18.00974. Epub 2019 Aug 7. Plant Cell. 2019. PMID: 31391256 Free PMC article.
-
Basic Helix-Loop-Helix (bHLH) Transcription Factors Regulate a Wide Range of Functions in Arabidopsis.Int J Mol Sci. 2021 Jul 1;22(13):7152. doi: 10.3390/ijms22137152. Int J Mol Sci. 2021. PMID: 34281206 Free PMC article. Review.
-
The Dof family of plant transcription factors.Trends Plant Sci. 2002 Dec;7(12):555-60. doi: 10.1016/s1360-1385(02)02362-2. Trends Plant Sci. 2002. PMID: 12475498 Review.
Cited by
-
Conserved language of plant gene regulation.Plant Physiol. 2025 Jul 3;198(3):kiaf263. doi: 10.1093/plphys/kiaf263. Plant Physiol. 2025. PMID: 40577606 Free PMC article. No abstract available.
References
-
- Appelhagen I, Jahns O, Bartelniewoehner L, Sagasser M, Weisshaar B, Stracke R. Leucoanthocyanidin Dioxygenase in Arabidopsis thaliana: characterization of mutant alleles and regulation by MYB–BHLH–TTG1 transcription factor complexes. Gene. 2011:484(1–2):61–68. 10.1016/j.gene.2011.05.031 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

