PTMA controls cardiomyocyte proliferation and cardiac repair by enhancing STAT3 acetylation
- PMID: 40408476
- PMCID: PMC12101487
- DOI: 10.1126/sciadv.adt9446
PTMA controls cardiomyocyte proliferation and cardiac repair by enhancing STAT3 acetylation
Abstract
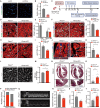
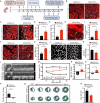
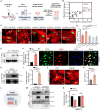
The adult mammalian heart has limited regenerative capacity due to the low proliferative ability of cardiomyocytes, whereas embryonic cardiomyocytes exhibit robust proliferative potential. Using single-cell RNA sequencing of embryonic hearts, we identified prothymosin α (PTMA) as a key factor driving cardiomyocyte proliferation. Overexpression of PTMA in primary mouse and rat cardiomyocytes significantly promoted cardiomyocyte proliferation and similarly enhanced proliferation in human iPSC-derived cardiomyocytes. Conditional knockout of Ptma in cardiomyocytes impaired neonatal heart regeneration. AAV9-mediated overexpression of Ptma extended the neonatal proliferative window and showed therapeutic promise for enhancing adult heart regeneration. Mechanistically, PTMA interacted with MBD3, inhibiting its deacetylation activity within the MBD3/HDAC1 NuRD complex. This inhibition increased STAT3 acetylation, which positively regulated STAT3 phosphorylation and activation of its target genes. These findings establish PTMA as a critical regulator of heart regeneration and suggest its potential as a therapeutic target for ischemic myocardial injury.
Figures







References
-
- Zhu F., Meng Q., Yu Y., Shao L., Shen Z., Adult cardiomyocyte proliferation: A new insight for myocardial infarction therapy. J. Cardiovasc. Transl. Res. 14, 457–466 (2021). - PubMed
-
- Konstam M. A., Kramer D. G., Patel A. R., Maron M. S., Udelson J. E., Left ventricular remodeling in heart failure: Current concepts in clinical significance and assessment. JACC Cardiovasc. Imaging 4, 98–108 (2011). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

