Mechanical signal modulates prostate cancer immune escape by USP8-mediated ubiquitination-dependent degradation of PD-L1 and MHC-1
- PMID: 40410130
- PMCID: PMC12102395
- DOI: 10.1038/s41419-025-07736-4
Mechanical signal modulates prostate cancer immune escape by USP8-mediated ubiquitination-dependent degradation of PD-L1 and MHC-1
Abstract
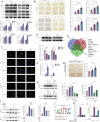
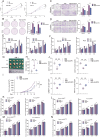
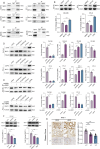
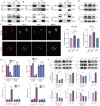
The tumor environment of prostate cancer (PCa) tissues of high Gleason score has been proved to be more immune suppressive and has higher extracellular matrix (ECM) stiffness, but whether ECM mechanical stiffness is the cause of higher ability of invasiveness and immune escape of PCa with high Gleason score remains uncertain. In this study, we showed that higher polyacrylamide hydrogels (PAAG) stiffness resulted in the progression and immune escape of PCa via integrin β1/FAK/YAP axis. The translocation of YAP into cell nucleus to bind to TEAD2 promoted the transcriptional activation of USP8. NBR1 could be ubiquitinated, and then degraded, via interacting with P62/SQSTM1 and through autophagy-lysosome pathway. Increased expression of USP8 promoted the abundance of NBR1 via K63-linked de-ubiquitination and PD-L1 via K48-linked de-ubiquitination in response to high PAAG stiffness. NBR1-mediated selective autophagy accelerated the degradation of MHC-1 of PCa. The USP8 inhibitor presented a potential application value in sensitizing immunotherapy of PCa. Taken together, we identified a USP8-mediated de-ubiquitination mechanism that involves in the process of high PAAG stiffness-mediated high expression of PD-L1 and low expression of MHC-1 of PCa cells, which provided a rationale of immunotherapy sensitization of PCa via USP8 inhibition.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethical approval and consent to participate: All experiments were performed according to the written confirmation that this study was in accordance with relevant institutional guidelines and regulations of Fujian Medical University and national guidelines and regulations of China. All animal experiments were approved by the Experimental Animal Ethics Committee of Fujian Medical University (Approved No. of ethic committee: IACUC FJMU 2023-Y-0722). Consent for publication: Informed consent for publication of the manuscript was obtained from all authors.
Figures





Similar articles
-
NDR1 mediates PD-L1 deubiquitination to promote prostate cancer immune escape via USP10.Cell Commun Signal. 2024 Sep 3;22(1):429. doi: 10.1186/s12964-024-01805-5. Cell Commun Signal. 2024. PMID: 39227807 Free PMC article.
-
Molecular mechanism of lncRNA SNHG12 in immune escape of non-small cell lung cancer through the HuR/PD-L1/USP8 axis.Cell Mol Biol Lett. 2022 Jun 3;27(1):43. doi: 10.1186/s11658-022-00343-7. Cell Mol Biol Lett. 2022. PMID: 35658874 Free PMC article.
-
USP8 inhibition reshapes an inflamed tumor microenvironment that potentiates the immunotherapy.Nat Commun. 2022 Mar 31;13(1):1700. doi: 10.1038/s41467-022-29401-6. Nat Commun. 2022. PMID: 35361799 Free PMC article.
-
What Do We Have to Know about PD-L1 Expression in Prostate Cancer? A Systematic Literature Review. Part 3: PD-L1, Intracellular Signaling Pathways and Tumor Microenvironment.Int J Mol Sci. 2021 Nov 15;22(22):12330. doi: 10.3390/ijms222212330. Int J Mol Sci. 2021. PMID: 34830209 Free PMC article.
-
New horizons in the mechanisms and therapeutic strategies for PD-L1 protein degradation in cancer.Biochim Biophys Acta Rev Cancer. 2024 Sep;1879(5):189152. doi: 10.1016/j.bbcan.2024.189152. Epub 2024 Jul 9. Biochim Biophys Acta Rev Cancer. 2024. PMID: 38992509 Review.
References
-
- Miller KD, Nogueira L, Devasia T, Mariotto AB, Yabroff KR, Jemal A, et al. Cancer treatment and survivorship statistics, 2022. CA Cancer J Clin. 2022;72:409–36. - PubMed
-
- O’Donnell JS, Teng MWL, Smyth MJ. Cancer immunoediting and resistance to T cell-based immunotherapy. Nat Rev Clin Oncol. 2019;16:151–67. - PubMed
-
- Achard V, Putora PM, Omlin A, Zilli T, Fischer S. Metastatic prostate cancer: treatment options. Oncology. 2022;100:48–59. - PubMed
-
- Adorno Febles VR, Hao Y, Ahsan A, Wu J, Qian Y, Zhong H, et al. Single-cell analysis of localized prostate cancer patients links high Gleason score with an immunosuppressive profile. Prostate. 2023;83:840–9. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

