Association of distinct microbial and metabolic signatures with microscopic colitis
- PMID: 40410138
- PMCID: PMC12102337
- DOI: 10.1038/s41467-025-59566-9
Association of distinct microbial and metabolic signatures with microscopic colitis
Abstract
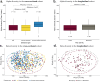
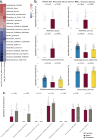
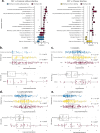
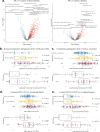
Microscopic colitis (MC) is a chronic inflammatory disease of the large intestine that primarily affects older adults and presents with chronic diarrhea. The etiology is unknown and there are currently no FDA approved medications or biomarkers for treatment or monitoring of the disease. Emerging evidence have implicated the gut microbiome and metabolome disturbances in MC pathogenesis. We conduct a comprehensive analysis of gut microbial and metabolic changes in a cohort of 683 participants, including 131 patients with active MC, 159 with chronic diarrhea, and 393 age- and sex-matched controls without diarrhea. Stool microbiome and metabolome are profiled using whole-genome shotgun metagenomic sequencing and ultra-high performance liquid chromatography-mass spectrometry, respectively. Compared to controls, eight microbial species including pro-inflammatory oral-typical Veillonella dispar and Haemophilus parainfluenzae, and 11 species, including anti-inflammatory Blautia glucerasea and Bacteroides stercoris are enriched and depleted in MC, respectively. Pro-inflammatory metabolites, including lactosylceramides, ceramides, lysophospholipids, and lysoplasmalogens, are enriched in active MC. Multi-omics analyses reveal robust associations between microbial species, metabolic pathways, and metabolites, suggesting concordant disruptions in MC. Here, we show distinct shifts in gut microbiome and metabolome in MC that can inform the development of non-invasive biomarkers and novel therapeutics.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: H.Kh. has served on clinical advisory board for Cylinder and has received consulting fees from Aditium Bio for work unrelated to the topic. K.S. has served as a consultant to Ardelyx, Gemelli Biotech, Laborie, Mahana, Salix, and Takeda and has received research support from Ardelyx and ReStalsis for work unrelated to the topic. Other authors have no competing interests to disclose. Ethical approval: The study was approved by Partners Human Research Committee and the Institutional Review Board of Mass General Brigham (Protocol # 2015P001333 and # 2015P000275).
Figures


Update of
-
Composition and Function of the Gut Microbiome in Microscopic Colitis.medRxiv [Preprint]. 2024 Oct 29:2024.10.28.24316293. doi: 10.1101/2024.10.28.24316293. medRxiv. 2024. Update in: Nat Commun. 2025 May 23;16(1):4644. doi: 10.1038/s41467-025-59566-9. PMID: 39574841 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
- P30 DK043351/DK/NIDDK NIH HHS/United States
- R01 AG068390/AG/NIA NIH HHS/United States
- P30DK043351/U.S. Department of Health & Human Services | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (National Institute of Diabetes & Digestive & Kidney Diseases)
- R01AG068390/U.S. Department of Health & Human Services | NIH | National Institute on Aging (U.S. National Institute on Aging)
LinkOut - more resources
Full Text Sources

