The health impacts and genetic architecture of food liking in cardio-metabolic diseases
- PMID: 40410146
- PMCID: PMC12102323
- DOI: 10.1038/s41467-025-59945-2
The health impacts and genetic architecture of food liking in cardio-metabolic diseases
Abstract
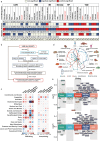
We evaluated temporal and genetic relationships between 176 food-liking-traits and cardio-metabolic diseases using data from the UK Biobank (N = 182,087) for observational analyses and summary-level GWAS data from FinnGen and other consortia (N = 406,565-977,323) for genetic analyses. Integrating observational and genetic results, we identified two detrimental food-liking-traits (bacon and diet-fizzy-drinks) and three protective food-liking-traits (broccoli, pizza, and lentils/beans). These food-liking-traits are associated with habitual food intake and influence cardio-metabolic proteins and biological processes. Notably, we found three genetic links: diet-fizzy-drinks with heart-failure, bacon with type-2-diabetes, and lentils/beans with type-2-diabetes, identifying 54 pleiotropic single-nucleotide-variants, impacting both phenotypes. Our data show the diet-fizzy-drinks and heart-failure link maybe not direct, as diet-fizzy-drinks liking correlates with sweet food consumption and shares variants linked to BMI, adiposity, platelet count and cardio-metabolic traits. The pleiotropic single-nucleotide-variants map to 251 tissue-specific genes, with four showing high druggability potential, highlighting personalized dietary strategies for cardio-metabolic diseases.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures







References
MeSH terms
Grants and funding
- 82173498/National Natural Science Foundation of China (National Science Foundation of China)
- 82204017/National Natural Science Foundation of China (National Science Foundation of China)
- 62472131/National Natural Science Foundation of China (National Science Foundation of China)
- 62172131/National Natural Science Foundation of China (National Science Foundation of China)
LinkOut - more resources
Full Text Sources
Medical

