Antibody-dependent enhancement of SARS-CoV-2, the impact of variants and vaccination
- PMID: 40411306
- PMCID: PMC12118418
- DOI: 10.1080/21645515.2025.2505356
Antibody-dependent enhancement of SARS-CoV-2, the impact of variants and vaccination
Abstract
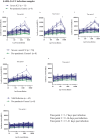
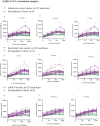
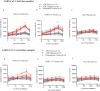
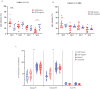
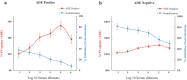
This study characterized antibody-dependent enhancement (ADE) in serum samples from individuals exposed to SARS-CoV-2 via infection or vaccination and evaluated its association with SARS-CoV-2 variants (Wuhan and Omicron), MERS-CoV, and NL63. ADE assays were performed on sera from SARS-CoV-2-infected patients (n = 210) with varying disease severity and vaccinated individuals (n = 225) who received adenovirus vector, inactivated virus or mRNA vaccines. ADE was assessed using pseudoviruses (PVs) in BHK cells expressing FcgRIIa. Neutralizing antibody levels, total IgG, IgG subclasses, and complement activation were analyzed using ELISA and neutralization assays. ADE was observed in 6.2% of infection samples (primarily severe cases) and 5.3% of vaccinated samples (adenovirus-vector and inactivated virus groups). ADE-positive samples showed reduced neutralizing activity, while total IgG and IgG subclasses did not differ significantly between ADE-positive and negative samples. Complement activation was elevated in severe cases but did not correlate clearly with ADE. Notably, MERS-CoV PV induced ADE in a subset of infected samples, but no ADE was detected for NL63. ADE was observed in SARS-CoV-2-infected individuals, particularly in severe cases, and in those vaccinated with adenovirus-vector and inactivated virus vaccines, but not with mRNA vaccines. Cross-reactivity leading to ADE was detected for MERS-CoV but not for NL63. ADE was associated with reduced neutralizing antibody activity and elevated complement activation in severe infections, though the specific role of complement in ADE remains unclear. These findings highlight the need to investigate the mechanisms underlying ADE and its implications for vaccine design and post-infection immunity against respiratory viruses.
Keywords: ADE; SARS-CoV2; Virology; immunology; variants.
Conflict of interest statement
No potential conflicts of interest are reported by the authors(s).
Figures
Similar articles
-
Antibody-dependent enhancement (ADE) of SARS-CoV-2 in patients exposed to MERS-CoV and SARS-CoV-2 antigens.J Med Virol. 2024 May;96(5):e29628. doi: 10.1002/jmv.29628. J Med Virol. 2024. PMID: 38682568
-
Cross-Reactive Antibody Responses to Coronaviruses Elicited by SARS-CoV-2 Infection or Vaccination.Influenza Other Respir Viruses. 2024 May;18(5):e13309. doi: 10.1111/irv.13309. Influenza Other Respir Viruses. 2024. PMID: 38725111 Free PMC article.
-
Diversified humoral immunity and impacts of booster vaccines: SARS-CoV-2 antibody profile and Omicron BA.2 neutralization before and after first or second boosters.Microbiol Spectr. 2024 Oct 3;12(10):e0060524. doi: 10.1128/spectrum.00605-24. Epub 2024 Aug 20. Microbiol Spectr. 2024. PMID: 39162540 Free PMC article.
-
A 10 Year Long-Lived Cellular and Humoral MERS-CoV Immunity Cross-Recognizing the Wild-Type and Variants of SARS-CoV-2: A Potential One-Way MERS-CoV Cross-Protection Toward a Pan-Coronavirus Vaccine.J Med Virol. 2025 Jan;97(1):e70071. doi: 10.1002/jmv.70071. J Med Virol. 2025. PMID: 39822038 Free PMC article.
-
Role of antibody-dependent enhancement (ADE) in the virulence of SARS-CoV-2 and its mitigation strategies for the development of vaccines and immunotherapies to counter COVID-19.Hum Vaccin Immunother. 2020 Dec 1;16(12):3055-3060. doi: 10.1080/21645515.2020.1796425. Epub 2020 Aug 26. Hum Vaccin Immunother. 2020. PMID: 32845733 Free PMC article. Review.
References
-
- Yip MS, Leung HL, Li PH, Cheung CY, Dutry I, Li D, Daëron M, Bruzzone R, Peiris JS, Jaume M. Antibody-dependent enhancement of SARS coronavirus infection and its role in the pathogenesis of SARS. Hong Kong Med J. 2016;22(3 Suppl 4):25–31. - PubMed
-
- Wang TT, Sewatanon J, Memoli MJ, Wrammert J, Bournazos S, Bhaumik SK, Pinsky BA, Chokephaibulkit K, Onlamoon N, Pattanapanyasat K, et al. IgG antibodies to dengue enhanced for FcγRIIIA binding determine disease severity. Science. 2017;355(6323):395–398. doi: 10.1126/science.aai8128. - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
