Synthetic antibodies targeting EphA2 induce diverse signaling-competent clusters with differential activation
- PMID: 40411427
- PMCID: PMC12102760
- DOI: 10.1002/pro.70145
Synthetic antibodies targeting EphA2 induce diverse signaling-competent clusters with differential activation
Abstract
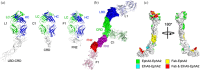
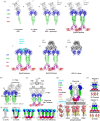
The receptor tyrosine kinase EphA2 interacts with ephrin (Efn) ligands to mediate bi-directional signals that drive cellular sorting processes during tissue development. In the context of various cancers, EphA2 can also drive invasive metastatic disease and represents an important target for cancer therapeutics. Natural Efn ligands sterically seed intertwined EphA2 clusters capable of recruiting intracellular kinases to mediate trans-phosphorylation. Synthetic proteins, such as antibodies (Abs), can mimic Efn ligands to trigger EphA2 signaling, leading to receptor internalization and degradation, and enabling intracellular delivery of conjugated drugs. Furthermore, Abs are capable of recruiting EphA2 into clusters distinct from those seeded by Efn. We developed three synthetic Abs targeting distinct EphA2 domains and determined the paratope valency necessary for agonist or antagonist properties of each of the three epitopes. Structural modeling of monovalent Fabs in complex with EphA2 elucidated competitive and non-competitive mechanisms of inhibition of EphA2 canonical signaling. Likewise, modeling of clusters induced by bivalent IgGs elucidated multiple signaling-competent EphA2 clusters capable of triggering a continuum of signaling strengths and provided insights into the requirement for multimerization of EphA2 to trigger phosphorylation. Our study shows how different agonist clusters lead to distinct kinase recruitment efficiencies to modify phosphotyrosine signal strength, and provides a panel of anti-EphA2 Abs as reagents for the development of therapeutics.
Keywords: Efn; Eph; EphA2; antibody; mechanism; phage display; receptor clustering; receptor tyrosine kinase; signaling.
© 2025 The Author(s). Protein Science published by Wiley Periodicals LLC on behalf of The Protein Society.
Conflict of interest statement
The authors declare no conflicting interests.
Figures






References
-
- Adams JJ, Nelson B, Sidhu SS. Recombinant genetic libraries and human monoclonal antibodies. Methods Mol Biol. 2014;1060:149–170. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

