Subclonal emergence of polycythemia vera, chronic myelomonocytic leukemia, and chronic myeloid leukemia
- PMID: 40411597
- PMCID: PMC12334534
- DOI: 10.1007/s00277-025-06417-8
Subclonal emergence of polycythemia vera, chronic myelomonocytic leukemia, and chronic myeloid leukemia
Abstract
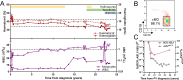
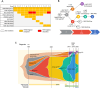
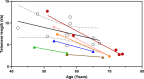
The present longitudinal study reports a unique patient followed over almost three decades who sequentially developed polycythemia vera, chronic myelomonocytic leukemia, and chronic myeloid leukemia. The patient received successive hydroxyurea, ruxolitinib, and a combination of ruxolitinib and nilotinib. The clonal architecture dynamic was reconstructed using targeted high throughput asymmetric capture sequencing, allowing detection and quantification of mutations in 43 myeloid genes and BCR::ABL1 fusion in multiple bone marrow or peripheral blood samples and in single cell-derived colonies obtained from bone marrow colony-forming cell assays. This analysis has uncovered an unexpected subclonal link between three myeloid malignancies, all stemming from a DNMT3A/TET2 double mutant clone. Over a period of more than 30 years, this clone underwent major telomere shortening. However, a striking sustained major molecular response of the terminal dominant clone carrying all driver mutations was achieved by combination therapy with nilotinib and ruxolitinib. The remaining clone driving both polycythemia and chronic myelomonocytic leukemia remained unaffected and evolved to myelofibrosis and proliferative CMML.
Keywords: Chronic myeloid leukemia; Chronic myelomonocytic leukemia; Co-occurrence; Dual targeting; Dual targeting myeloproliferative neoplasm; Polycythemia vera; Ruxolitinib; Telomere; Tyrosine kinase inhibitor; aCAP-Seq.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This case report complied with French regulations and was approved by the Henri Mondor. Institutional Review Board (No. 00011558). The study methodologies conformed to the standards set by the Declaration of Helsinki. All patient data were anonymized and de-identified before analysis, and informed consent was obtained from all patients. Consent for publication: All authors have agreed with the content of the manuscript for publication. Competing interests: IS is a speaker for Novartis and Incyte. IS is a co-inventor of a know-how licensed by AP-HP to Agilent Technologies. OWB has a patent issued relevant to this work. Other authors declare no conflict of interest.
Figures
References
-
- Gricourt G, Tran Quang V, Cayuela J-M et al (2022) Fusion gene detection and quantification by asymmetric capture sequencing (aCAP-Seq). J Mol Diagn 24:1113–1127. 10.1016/j.jmoldx.2022.07.004 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

