N4-acetylcytidine modification of LncRNA GFOD1-AS1 promotes high glucose-induced dysfunction in human dermal microvascular endothelial cells through stabilization of DNMT1 protein
- PMID: 40411601
- PMCID: PMC12103335
- DOI: 10.1007/s10142-025-01617-x
N4-acetylcytidine modification of LncRNA GFOD1-AS1 promotes high glucose-induced dysfunction in human dermal microvascular endothelial cells through stabilization of DNMT1 protein
Abstract
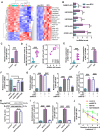
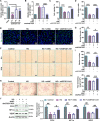
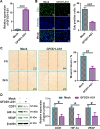
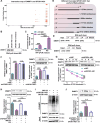
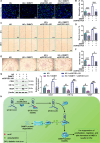
Emerging evidence supports that angiogenesis is essential for the wound healing of diabetic foot ulcer (DFU), and high glucose (HG)-induced dysfunction of human dermal microvascular endothelial cells is a key factor that hinders angiogenesis. However, the underlying mechanisms by which HG leads to the dysfunction of human dermal microvascular endothelial cells has not been fully elucidated. In the present investigation, we discovered a significant upregulation of the long non-coding RNA GFOD1-AS1(GFOD1-AS1) in the ulcer margin samples of patients with DFU and the HG-induced dysfunction model of human dermal microvascular endothelial cells, attributing its dysregulation to the stabilizing effect of NAT10-mediated ac4C modification, as corroborated by an integrated approach of data mining and experimental validation. Subsequently, a series of in vitro functional analyses showed that ectopic expression of GFOD1-AS1 promoted impaired function of human dermal microvascular endothelial cells. In contrast, knockdown of GFOD1-AS1 significantly alleviated the HG-induced functional impairment in human dermal microvascular endothelial cells, as indicated by the enhanced cell proliferation, migration, and tube formation. Mechanistically, GFOD1-AS1 directly interacts with DNA methyltransferase DNMT1 to block its ubiquitin-proteasome degradation, thereby enhancing the protein stability of DNMT1.This stability elevates DNMT1 protein expression, ultimately inducing HG-induced dysfunction in human dermal microvascular endothelial cells. In summary, our results reveal that GFOD1-AS1 serves as a potential therapeutic target for DFU, and highlight the critical role of the NAT10/GFOD1-AS1/DNMT1 axis in the dysfunction of human dermal microvascular endothelial cells in DFU.
Keywords: DNMT1; Diabetic foot ulcer; GFOD1-AS1; Human dermal microvascular endothelial cells; N4-acetylcytidine modification.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures
Similar articles
-
LncRNA A1BG-AS1 regulates the progress of diabetic foot ulcers via sponging miR-214-3p.Endocr J. 2025 Mar 3;72(3):295-306. doi: 10.1507/endocrj.EJ24-0440. Epub 2025 Jan 7. Endocr J. 2025. PMID: 39779214 Free PMC article.
-
Long Non-coding RNA SPAG5-AS1 Attenuates Diabetic Retinal Vascular Dysfunction by Inhibiting Human Retinal Microvascular Endothelial Cell Proliferation, Migration, and Tube Formation by Regulating the MicroRNA-1224-5p/IRS-1 Axis.Mol Biotechnol. 2023 Jun;65(6):904-912. doi: 10.1007/s12033-022-00572-3. Epub 2022 Nov 8. Mol Biotechnol. 2023. PMID: 36346578
-
lncRNA ANRIL accelerates wound healing in diabetic foot ulcers via modulating HIF1A/VEGFA signaling through interacting with FUS.J Gene Med. 2023 Feb;25(2):e3462. doi: 10.1002/jgm.3462. Epub 2022 Dec 4. J Gene Med. 2023. PMID: 36346049
-
LncRNA SAMD12-AS1 Promotes the Progression of Gastric Cancer via DNMT1/p53 Axis.Arch Med Res. 2021 Oct;52(7):683-691. doi: 10.1016/j.arcmed.2021.04.004. Epub 2021 May 4. Arch Med Res. 2021. PMID: 33962804
-
USP7-stabilised HIPK2 promotes high glucose-induced endothelial cell dysfunctions to accelerate diabetic foot ulcers.Arch Physiol Biochem. 2024 Dec;130(6):984-991. doi: 10.1080/13813455.2024.2376815. Epub 2024 Jul 27. Arch Physiol Biochem. 2024. PMID: 39066661
References
-
- Bian EB, Xiong ZG, Li J (2019) New advances of LncRNAs in liver fibrosis, with specific focus on lncRNA-miRNA interactions. J Cell Physiol 234:2194–2203 - PubMed
-
- Chen L, Shen S, Wang S (2023) LncRNA SNHG16 knockdown promotes diabetic foot ulcer wound healing via sponging MiR-31-5p. Tohoku J Exp Med - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

