Complete CD16A Deficiency and Defective NK Cell Function in a Man Living with HIV
- PMID: 40411624
- PMCID: PMC12103316
- DOI: 10.1007/s10875-025-01886-y
Complete CD16A Deficiency and Defective NK Cell Function in a Man Living with HIV
Abstract
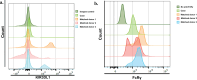
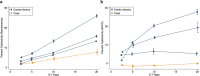
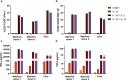
A man living with HIV was found to lack expression of CD16A on his natural killer (NK) cells and monocytes. Genetic analysis revealed compound heterozygous deletion of FCGR3A, the gene encoding CD16A. The case's NK cells showed: (a) no antibody-dependent cell-mediated cytotoxicity and very low spontaneous cytotoxicity; (b) an immature phenotype marked by high expression of CD94, CD2, NKG2A, and NKG2D, and low expression of KIR2DL2 and CD57; (c) no expression of KIR3DL1 and very low expression of FcRγ; and (d) normal cytokine production. The case's monocytes and DCs were similar phenotypically and functionally to those from the donors matched for HIV status, age, and percentage of NK cells in the peripheral blood. In contrast to previously reported people with CD16A deficiency, this man did not have a history of severe infections with herpes viruses, suggesting that other immune cells and/or immunoregulatory function of NK cells may compensate for deficiency of cytolytic NK cells.
Keywords: CD16A deficiency; Dendritic cell function; FCGR3A deletion; Monocyte function; NK cell function.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing Interests: The authors declare no competing interests.
Figures



References
-
- Bruhns P. Properties of mouse and human IgG receptors and their contribution to disease models. Blood. 2012;119:5640–9. - PubMed
-
- Nimmerjahn F, Ravetch JV. Fcγ receptors as regulators of immune responses. Nat Rev Immunol. 2008;8:34–47. - PubMed
-
- Guilliams M, Bruhns P, Saeys Y, Hammad H, Lambrecht BN. The function of Fcγ receptors in dendritic cells and macrophages. Nat Rev Immunol. 2014;14:94–108. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

