MePCE promotes homologous recombination through coordinating R-loop resolution at DNA double-stranded breaks
- PMID: 40411785
- PMCID: PMC12261951
- DOI: 10.1016/j.celrep.2025.115740
MePCE promotes homologous recombination through coordinating R-loop resolution at DNA double-stranded breaks
Abstract
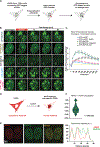
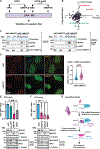
MePCE is a multifunctional protein that regulates the positive transcription elongation factor b (P-TEFb) partitioning between the nucleosol and chromatin. MePCE's role in sequestering P-TEFb in the nucleosol via the 7SK ribonuclear protein complex (RNPc) is clear, but its functions on chromatin remain obscure. We report that chromatin-associated MePCE interacts with R-loop processing and DNA repair factors. MePCE is recruited to DNA double-stranded breaks (DSBs), and MePCE depletion impairs DSB repair by homologous recombination (HR), decreases RAD51 loading, and enhances R-loop levels at AsiSI-induced DSBs at specific genomic locations. Besides decreasing specific R-loop processing factors and chromatin remodelers, MePCE depletion increases the interaction with R-loops of the other constitutive member of the 7SK RNPc, LARP7, which is degraded by BRCA1/BARD1 upon DSB. Overall, our results uncover dynamic regulation of the 7SK RNPc at DSBs during the DSB repair process and explain the recently observed synthetic lethality of MePCE and BRCA1 deficiency.
Keywords: 7SK; BRCA1; CP: Molecular biology; DNA repair; DRIP-MS; DRIP-seq; FACT; LARP7; MePCE; R-loops; homologous recombination.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
-
- Jeronimo C, Forget D, Bouchard A, Li Q, Chua G, Poitras C, Thérien C, Bergeron D, Bourassa S, Greenblatt J, et al. (2007). Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol. Cell 27, 262–274. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

