Gut microbiota mediates semaglutide attenuation of diabetes-associated cognitive decline
- PMID: 40413074
- PMCID: PMC12491809
- DOI: 10.1016/j.neurot.2025.e00615
Gut microbiota mediates semaglutide attenuation of diabetes-associated cognitive decline
Abstract
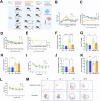
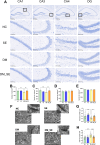
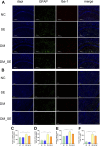
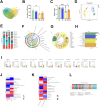
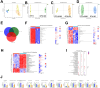
Diabetes-associated cognitive decline (DACD), characterized by cognitive impairment, is a serious complication of diabetes mellitus (DM). Research has shown that semaglutide, a novel glucagon-like peptide-1 receptor agonist, has neurotrophic and neuroprotective properties. However, a comprehensive understanding of the specific effects and underlying mechanisms of semaglutide treatment in patients with DACD remains lacking. In this study, we evaluated the potential of semaglutide to alleviate DACD in mice with DM. Eight-week-old mice fed a high-fat diet with streptozotocin-induced DM were subcutaneously injected with semaglutide (30 nmol/kg qd) for 12 weeks. Semaglutide administration significantly alleviated cognitive impairment, inhibited hippocampal neuron loss, improved the hippocampal synaptic ultrastructure, and effectively mitigated neuroinflammation. Furthermore, semaglutide treatment increased the relative abundances of g_Alistipes, g_norank_f_Eubacterium_coprostanoligenes, g_Bacteroides, and g_Parabacteroides, while decreasing the relative abundances of g_ faecalibaculum, g_Colodertribacter, g_GCA-900066575, g_Erysipelatoclostridium, and g_norank_f_Lachnospiraceae. Semaglutide also induced alterations in fecal and serum metabolites, as well as transcriptomic changes in brain tissue, with significant common enrichment in neuroactive ligand-receptor interactions. Furthermore, strong correlations were observed among semaglutide-affected genes, metabolites, and microbiota, as assessed by correlation analysis and integrative modeling. In conclusion, these findings suggest a correlation between the protective effects of semaglutide against DACD and the microbiota-gut-brain axis.
Keywords: Cognitive impairment; Gut microbiota; Metabolome; Neuroactive ligand-receptor interactions; Semaglutide.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors have declared no competing interests.
Figures





References
-
- Saeedi P., Petersohn I., Salpea P., Malanda B., Karuranga S., Unwin N., et al. Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: results from the international diabetes federation diabetes atlas. Diabetes Res Clin Pract. 2019;157 9th edition. - PubMed
-
- Luo A., Xie Z., Wang Y., Wang X., Li S., Yan J., et al. Type 2 diabetes mellitus-associated cognitive dysfunction: advances in potential mechanisms and therapies. Neurosci Biobehav Rev. 2022;137 - PubMed
-
- Ye M., Liu Y., Wang F., Yang X., Yang X., Gao X., et al. Polysaccharide extracted from Sarcandra glabra residue attenuate cognitive impairment by regulating gut microbiota in diabetic mice. Int J Biol Macromol. 2024;270(Pt 1) - PubMed
-
- Morais L.H., Schreiber H.L., 4th, Mazmanian S.K. The gut microbiota-brain axis in behaviour and brain disorders. Nat Rev Microbiol. 2021;19(4):241–255. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

